Binding peptides specific for the extracellular domain of ErbB2 and uses therefor
a technology of erbb2 and erbb2, which is applied in the field of peptides and peptidomimetics, can solve the problems of poor prognosis, lack of specificity and associated toxicities to normal tissues, and achieve the effects of improving pharmacokinetic properties and facilitating detection, prevention and treatmen
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Identification of Peptide Phage from Random and Biased Peptide Phage Libraries
[0252] Clones were identified by screening random as well as biased peptide phage library. Peptides identified by screening random libraries include those having the following sequences:
CB022701-20DTDMCWWWSREFGWECAGAG(SEQ ID NO: 37)(E-20)CB051701-19SLALCLSEGVLLGADCRVLF(SREQ ID NO: 38)(C-19)CB051701-25WSSMCGDPTIADWLWCFSDA.(SEQ ID NO: 39)(C-25)
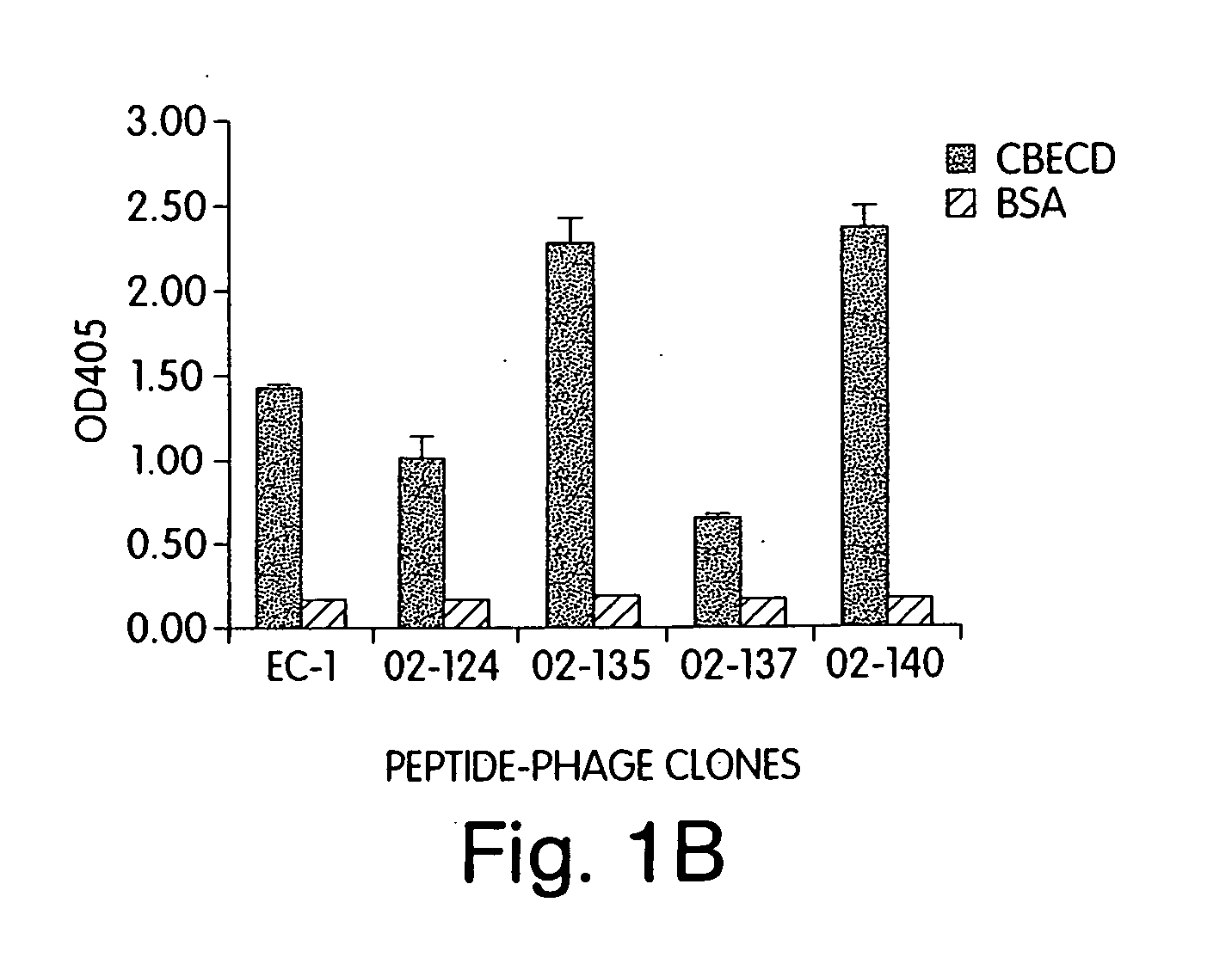
[0253] The following example describes the methodology used to identify four clones from four biased peptide phage libraries. The libraries were enriched for the sequence of the EC-1 phage clone (SEQ ID NO:1). The four libraries had the following design:
X4CLNPEESTWGFCRSAG,(SEQ ID NO: 29)WTGWCX5STWGFCRSAG,(SEQ ID NO: 30)WTGWCLNPEEX5CRSAG,(SEQ ID NO: 31)andWTGWCLNPEESTWGFCX4,(SEQ ID NO: 32)
where X=any amino acid.
[0254] The peptides so identified had the following sequences:
02-124WTGWCLNPEESTWGFCRSAG(SEQ ID NO: 33)02-137WTGWCLSPEESTWGFCRSAG(SEQ ID NO: 34)02-140WT...
example 2
Binding of Peptide Phage to Membrane Lysates from ErbB2 Expressing Cells
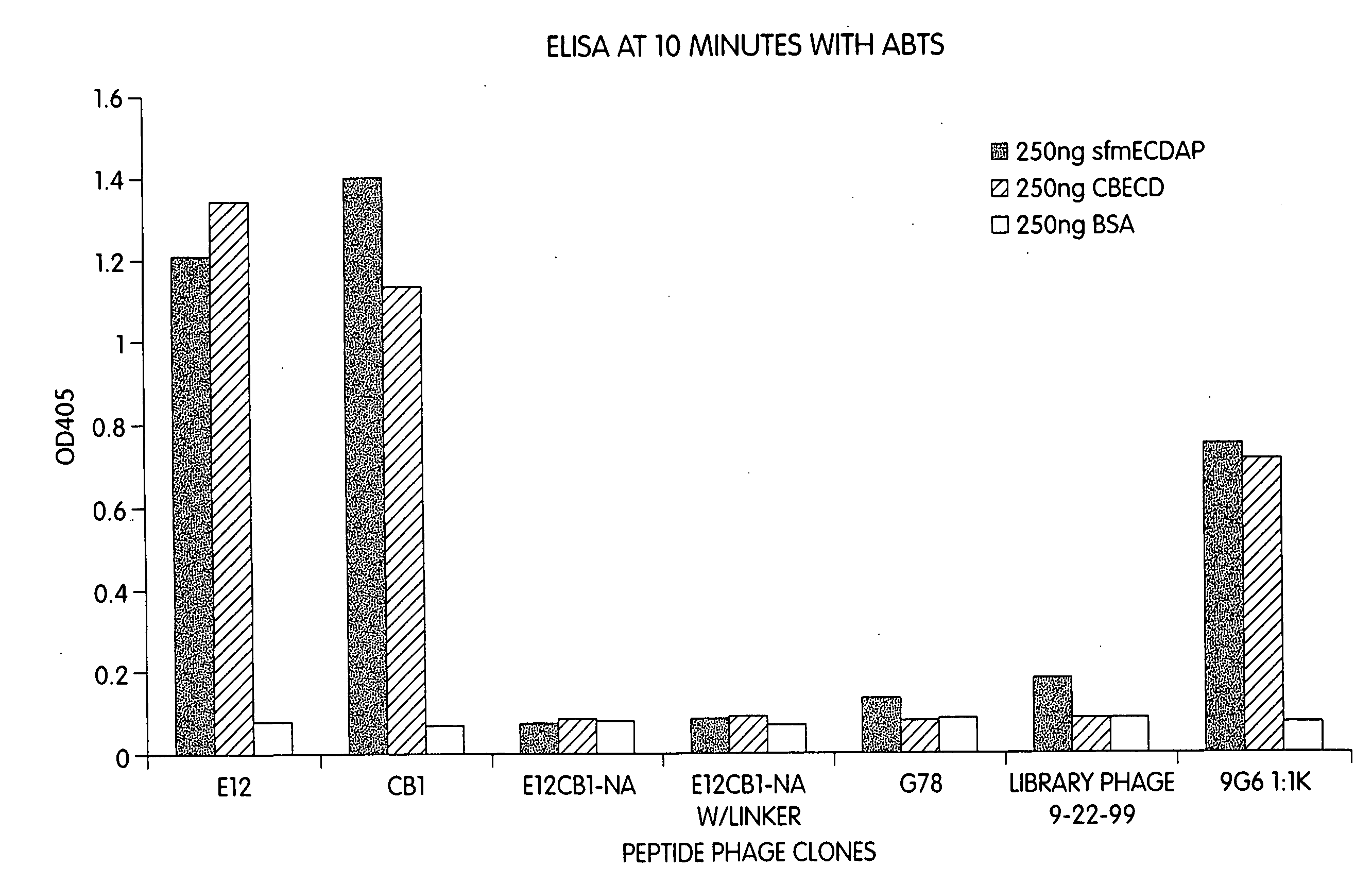
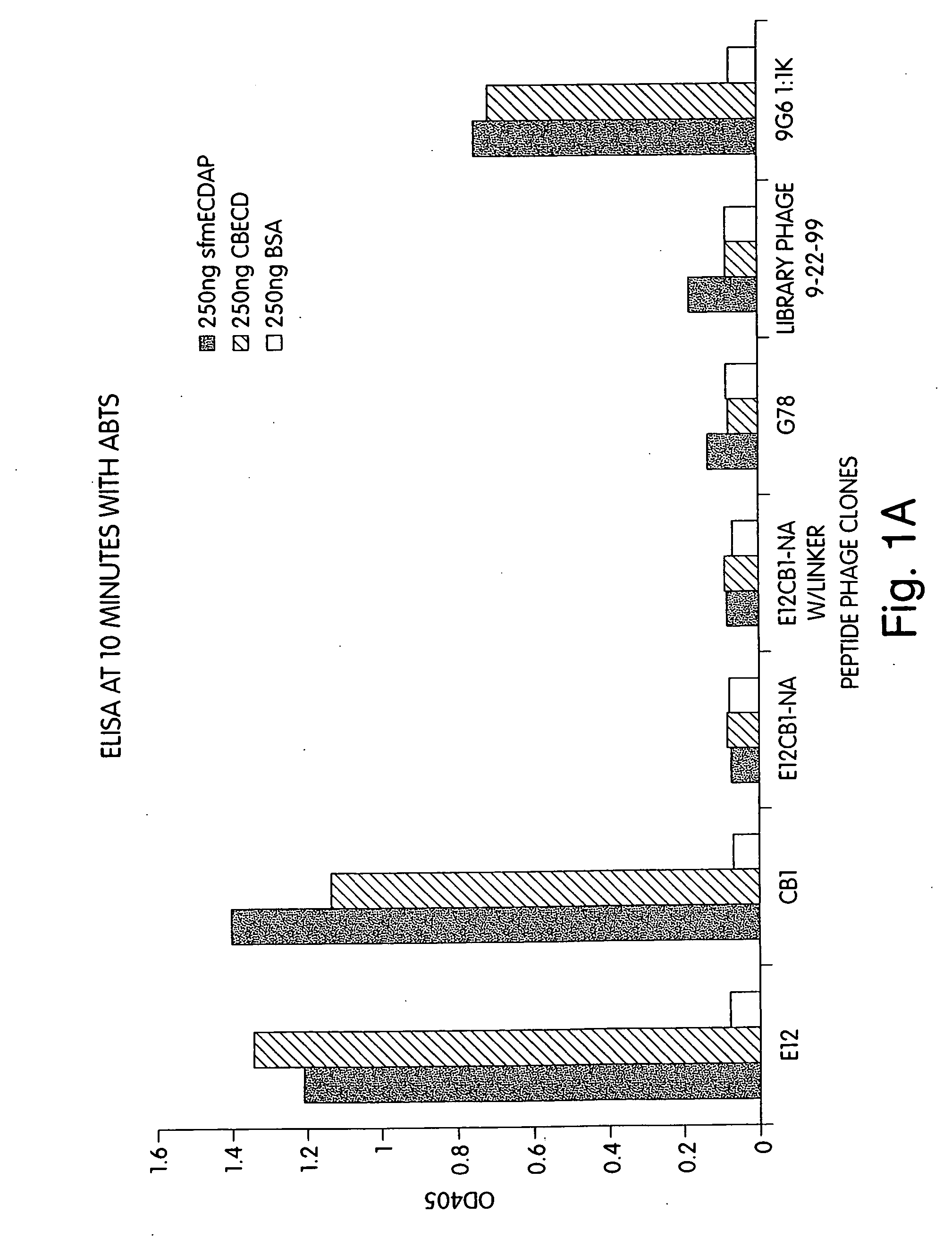
[0258] The above identified peptide phage specifically bind to native, intact ErbB2 extracted from the membranes of human breast cancer cells that overexpress ErbB2. Plates were coated with 1 μg of an antibody that binds to the intracellular domain of ErbB2, followed by 100 μl of solubilized membrane preparations from two different breast cancer cell lines that overexpress ErbB2 (BT474 and SKBR3), and probed with CB1 peptide-phage, control library phage, and 9G6 anti-ErbB2. MCF7 cells serve as a (relative) negative target control, as MCF7 breast cancer cells express only a low level of ErbB2. (See FIG. 2A.) The ELISA data indicates that the EBP gives a signal higher than the anti-ErbB2 antibody used as a positive control in the assay.
[0259] Peptide phage derived from the EC-1 biases library similarly bind specifically to membrane lysates from ErbB2 expressing SKBR3 cells but not to lysates from minimally expre...
example 3
Effect of Phagepeptide on ErbB2 Phosphorylation
[0261] ErbB2 overexpressing SKBR3 cells were treated with the EC-1 free peptide to determine its effect on ErbB2 activation. When ErbB2 is activated it becomes phosphorylated on specific tyrosine residues (i.e., pY1248 and pY877). This is turn triggers downstream signal transduction events, culminating in increased cellular proliferation. Viable SKBR3 cells were treated with 25 μM EC-1 peptide for 15 minutes. At 0.5, 2, 4, 8, 22 and 48 hours thereafter, cell lysates were prepared and run on a Western blot using phospho-specific ErbB2 antibodies. (See FIG. 6A.) Densitometric analysis of the Western blots showed that EC-1 peptide inhibits 40% of the phosphorylation of residues pY1248 and pY877 after 0.5 hrs. The results demonstrate that 25 μM EC-1 inhibits the phosphorylation of ErbB2 for at least 8 hours after treatment. (See FIG. 6B.) The peptide had no effect on total ErbB2 expression.
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperatures | aaaaa | aaaaa |
| length | aaaaa | aaaaa |
| nucleic acid | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com