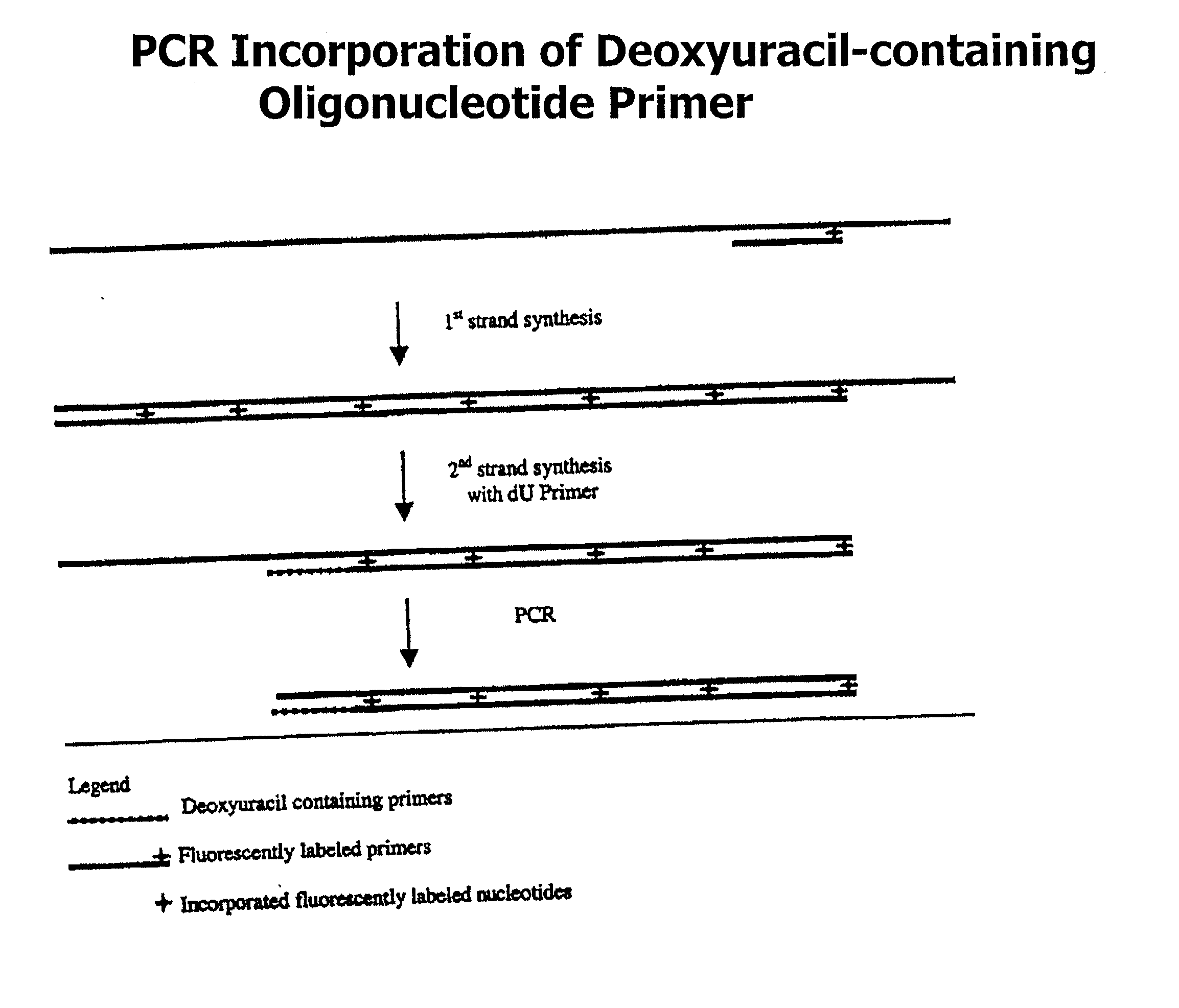
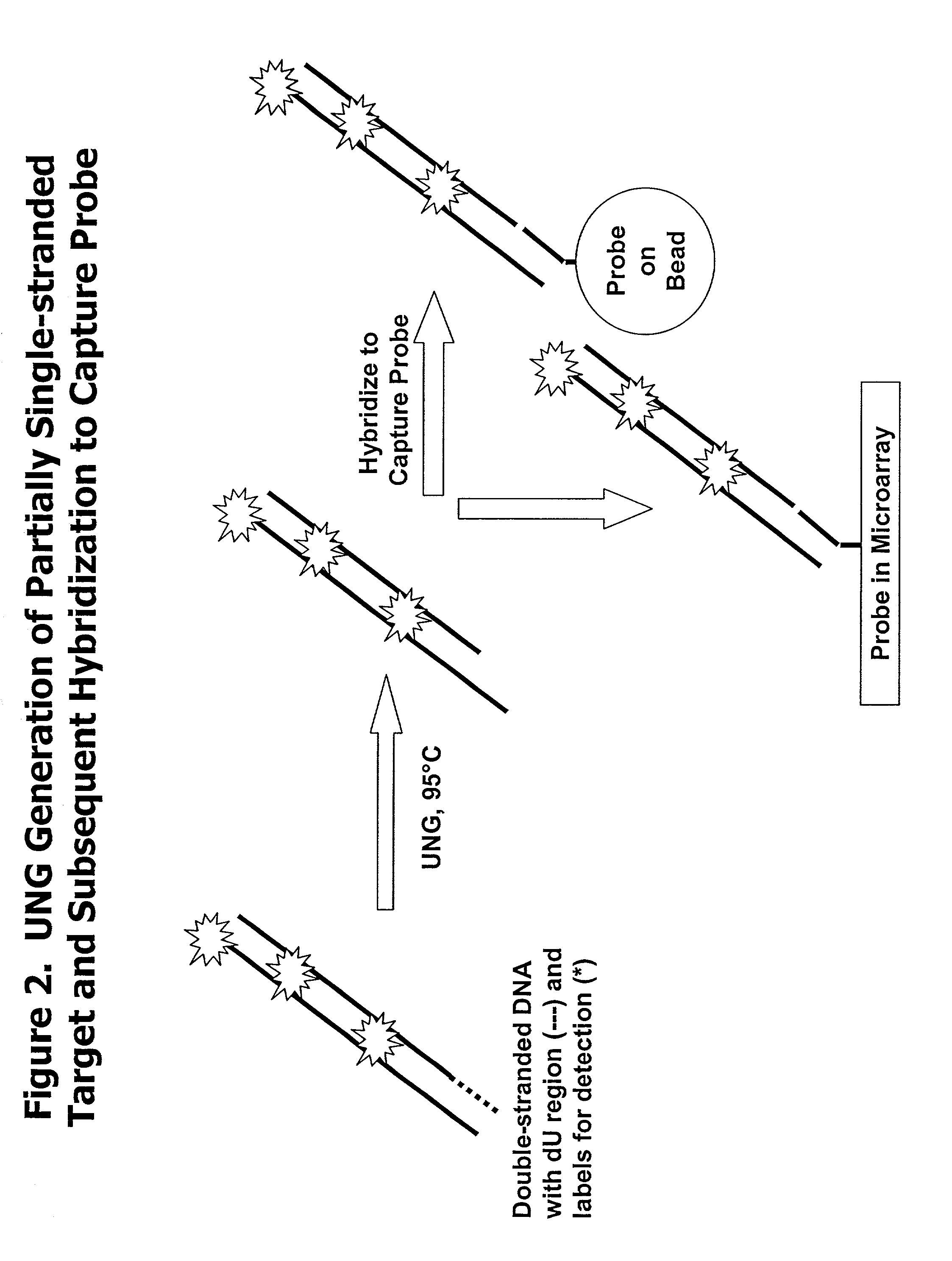
Partially double-stranded nucleic acids, methods of making, and use thereof
a nucleic acid, partially double-strand technology, applied in the direction of microbiological testing/measurement, biochemistry apparatus and processes, fermentation, etc., can solve the problems of inefficient and problematic strand separation steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
third embodiment
[0058] In one embodiment, P2 contains a detectable label, which is subsequently incorporated into the third region. In another embodiment a label is incorporated into P1 3' to any labile nucleotide. In a third embodiment, a detectable label is incorporated into the second region.
[0059] In another embodiment, the single-stranded region may be 10-50 nucleotides or more in length, or any integer value subsumed within that range.
[0060] In another aspect of the invention, there is provided a method for generating a partially double-stranded polynucleotide containing at least one single-stranded index sequence at a terminal end comprising:
[0061] preparing a first primer, Pa, comprising two regions:
[0062] a) a first region comprising a first index sequence containing at least one labile nucleotide wherein, the first region is not complementary to a target nucleic acid sequence; and
[0063] b) a second region that is specific for the target nucleic acid sequence;
[0064] preparing a second prim...
example 1
[0154] A sample containing the target nucleic acid was prepared as follows. RNA was isolated using the RNeasy kit (Qiagen, Valencia, Calif.) using the manufacturer's recommended conditions. This method is suitable for isolating up to 100 .mu.g of RNA, which is the approximate binding limit of the RNeasy mini spin column. All buffers mentioned below are provided with the RNeasy kit. The Buffer RLT was warmed to dissolve any precipitate, and then .beta.-mercapto-ethanol ("BME") (10 .mu.l per 1 ml of Buffer RLT) was added before use. Four volumes of 100% EtOH also was added to Buffer RPE before initial use. The sample (lysed and digested cells or tissue that is deproteinated and delipidated) was adjusted to 100 .mu.g / 100 .mu.l using RNase-free H.sub.2O. If the sample was more than 130 .mu.l, it was split into two tubes, and each was diluted to 100 .mu.l with RNAsecure. Samples were placed into a 1.5 ml tube(s), and 350 .mu.l of Buffer RLT was added, with mixing. Then ...
example 2
UNG Degradation of Forward Primer
[0158] The degradation of the dU-containing primer was assessed by performing a gel shift assay. Primers and probes were synthesized by standard synthesis procedures. A listing of primers and probes is given in FIG. 5. Double stranded amplicons were produced from a forward dU-containing primer and a reverse non-dU containing primer. The primers were incorporated into a double-stranded DNA molecule using a thermocycler according to the protocol in Table 1 and using RNA obtained by methods described in Example 1. The components used in the PCR amplification reaction can be found in Table 1. RT-PCR amplification was done according to the instructions of the thermocycler used, e.g. PERKIN ELMER GeneAmp PCR system. Optimization guidelines are provided with commercially available RT-PCR kits to adjust conditions to obtain the highest yield of RT-PCR product. Following amplification, samples were cooled to 4.degree. C.
1TABLE 1 RT-PCR Reaction Components Vol...
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| temperatures | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com