Polypeptides of alicyclibacillus
A Bacillus, alicyclic acid technology, applied in the direction of peptides, depsipeptides, bacteria, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0389] Example 1: Identification of functional polypeptides secreted by Alicyclobacillus DSM 15716
[0390] Genomic library construction
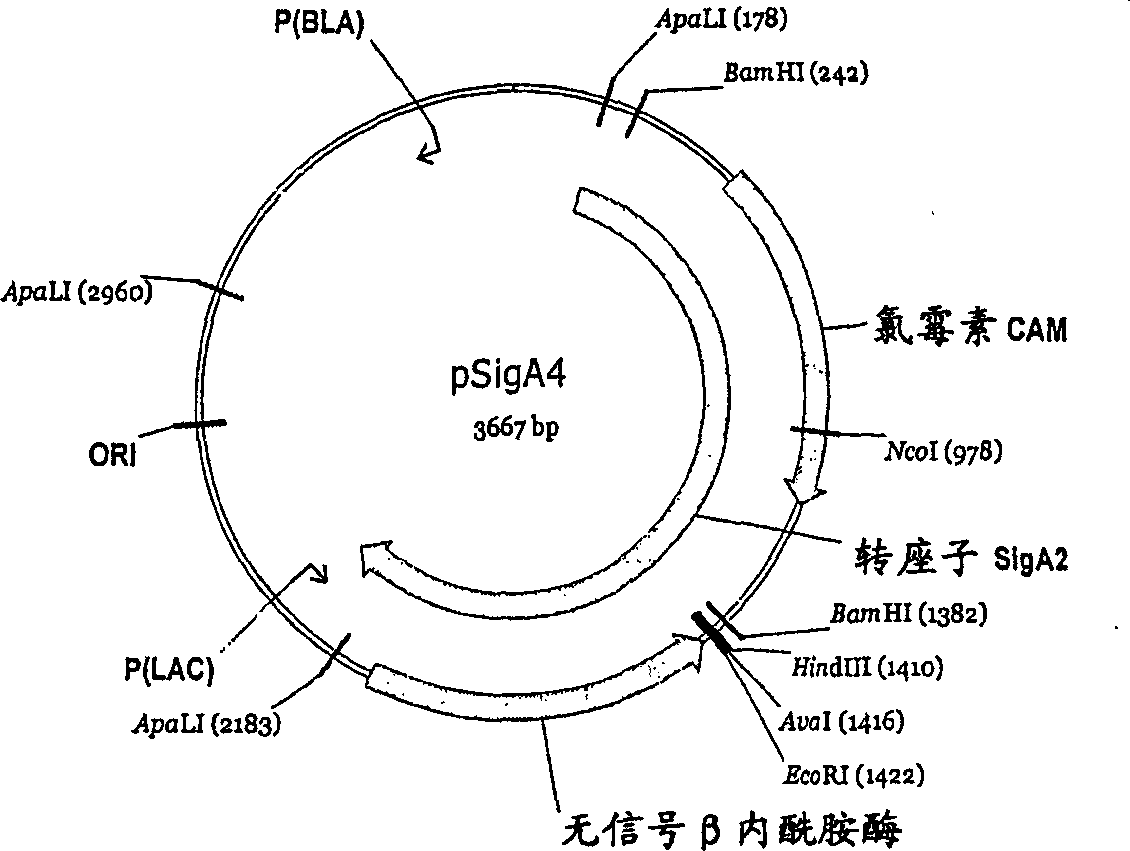
[0391] Chromosomal DNA of Alicyclobacillus DSM 15716 was prepared by using standard molecular biology techniques (Ausuble et al., 1995 "Current protocols in molecular biology", published by John Wiley and Sons). The prepared DNA was partially digested with Sau3A and separated on an agarose gel. Elute, pellet and resuspend the 3 to 8 kilobase fragments in an appropriate buffer.
[0392] By using Stratagene ZAP Express TM Predigested Vector Kit and StratageneZAP Express TM Predigested Gigapack Cloning kit (Bam HI predigested) (Stratagene Inc., USA) Genomic libraries were prepared according to the manufacturer's instructions / recommendations. The generated λZAP contained 38000 pfu, 10000 of which were selected for mass digestion. The resulting 70000 E. coli colonies were pooled and plasmids were prepared using the Qiagen Spin Mini prep k...
Embodiment 2
[0412] Example 2: Determination of function by homology
[0413] The functions of the polypeptides of SEQ ID NO: 26 to SEQ ID NO: 50 are annotated by comparison with gene or polypeptide sequences of known function. Polypeptides of the invention are compared to a list of most related sequences from public and internal contig databases. The BLASTX 2.Oa19MP-WashU [July 14, 1998] program was then used to compare the contigs (from which SEQ ID NO: 26 to SEQ ID NO: 50 were derived) to sequences available from standard public DNA and protein sequence databases. Careful analysis of sequence alignments of SEQ ID NO: 26 to SEQ ID NO: 40 with their most related functionally known sequences from other databases made it possible to predict the function of these polypeptides on the basis of the degree of amino acid identity. Even when the total amino acid identity is 40% (usually difficult to make a good prediction), we can predict SEQ ID NO: 26 to SEQ ID NO: 40 functions. When the amino...
Embodiment 3
[0414] Embodiment 3: Preparation of the polypeptide of SEQ ID NO: 26 to SEQ ID NO: 50
[0415] In order to prepare the polypeptide of SEQ ID NO: 26 to SEQ ID NO: 50, by combining the DNA encoding the open reading frame with a suitable host strain (such as Escherichia coli, Bacillus subtilis, Bacillus licheniformis or Bacillus clausii or lipid Bacillus cycloacidum derivatives) were fused to express the genes encoding these polypeptides contained in SEQ ID NO: 1 to SEQ ID NO: 25. Promoters can be inducible or constitutive. Any signal sequence of SEQ ID NO: 26 to SEQ ID NO: 50 can be exchanged for an appropriate signal peptide from another bacterium. Expression constructs can be part of a plasmid or linear DNA. It can be integrated into the chromosome of the host strain by recombination or it can be present in the host cell as a plasmid. Transformed cells carrying the gene of interest are then cultured in the desired volume in an appropriate medium. If an inducible promoter i...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com