Variants of enzymes of the alpha-amylase family
An amylase and family technology, applied in the field of enzyme variants, can solve problems such as unknown domain functions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0739] Error prone PCR conditions -
[0740] Optimal error-prone PCR mutagenesis conditions by using 10 different MnCl 2 The BC251cgt gene (SEQ ID 4) assay was amplified at the concentration. The PCR product was cloned into plasmid pDP66k-, transformed into E. coli and plated on starch-supplemented LB dishes. Only clones expressing the starch-degrading CGTase formed halos around the flora. The percentage of clones forming halos increased with MnCl 2 decreased with increasing concentration (Figure 3), indicating that MnCl 2 The higher the concentration, the more the number of inactivating mutations. Choose 0.2-0.3mM MnCl 2 concentration as the optimal error-prone PCR condition, in which the MnCl 2 Approximately 90% of the (mutant) CGTase clones obtained at the concentration retained starch degrading activity (Fig. 3).
Embodiment 2
[0742] Design variants for increased hydrolytic activity
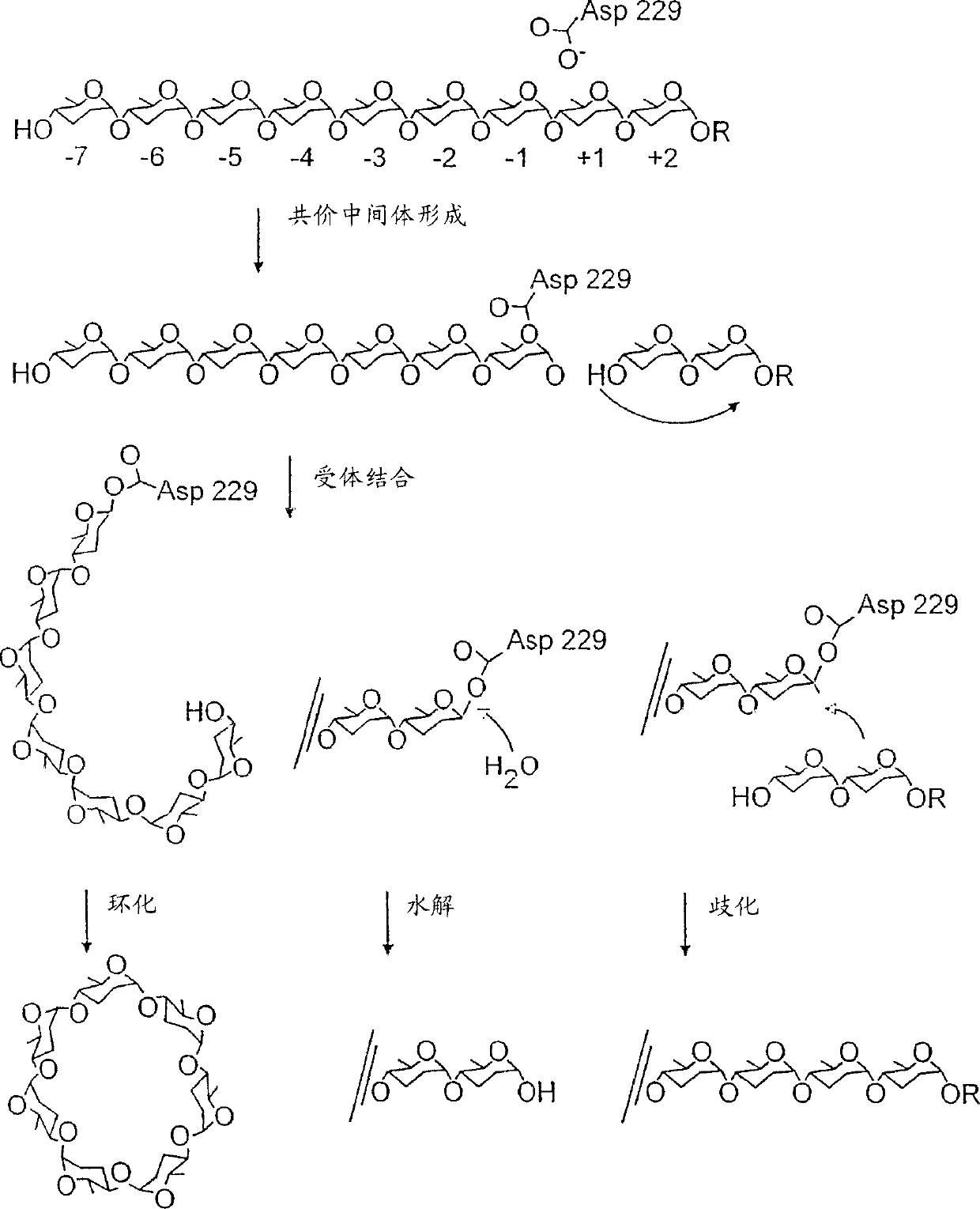
[0743] To select CGTase variants with increased hydrolytic activity, approximately 12,000 clones were analyzed for hydrolytic activity in a first round of mutagenesis and screening. MnCl used in PCR 2 Concentrations were 0.2 mM (6,000 clones) and 0.3 mM (6,000 clones). 22 clones had significantly increased hydrolytic activity. Plasmid DNA was isolated from each positive clone and subsequently used to prepare and purify the encoded mutant protein. All 22 mutant CGTases had higher hydrolytic activity and lower cyclization activity than wild-type CGTase. Some mutants even had higher hydrolytic activity than cyclization activity.
[0744] 0.25mM MnCl was used in PCR amplification 2 , the hydrolytic activity of CGTase was further enhanced in the second and third rounds of mutagenesis. Mutants 6 and 6-2 served as PCR templates in the second and third rounds of mutagenesis, respectively; these mutants had the highest ...
Embodiment 3
[0748] Mutations that enhance hydrolytic activity
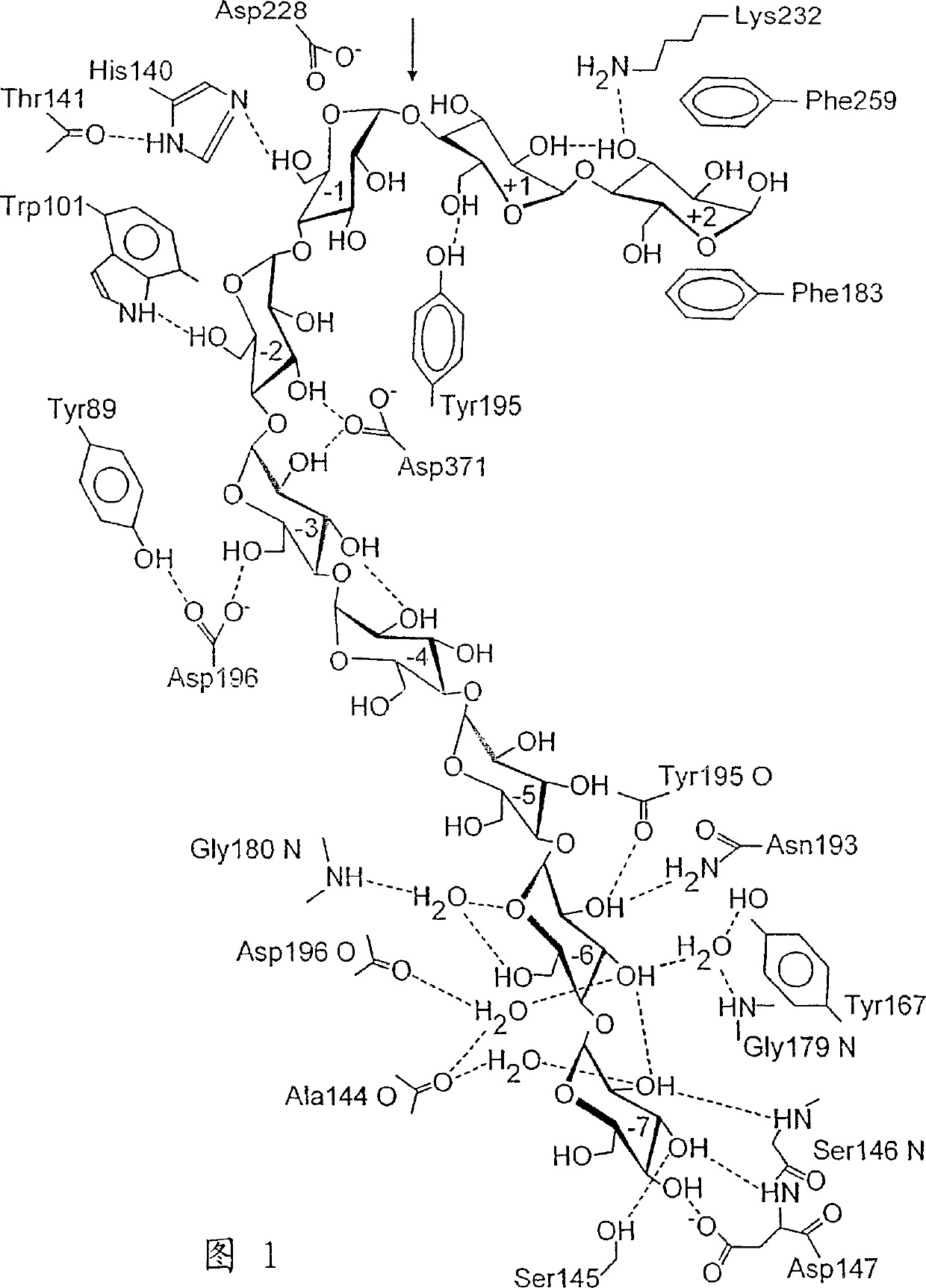
[0749] The 12 mutants showing the highest hydrolytic activity after the first round of mutagenesis had either the F259S (4 times) or the A230V (SEQ ID 1) (8 times) mutation, with or without the second mutation. A second round of mutagenesis resulted in the F21L mutation (3 times) and once a combination of A245T / A357T, in addition to the A230V / V660A mutation. After the third round of mutagenesis, mutations N8S, R47W, N94S and Q320L were identified, although N8S was only identified in combination with N94S. Among the mutations with significantly enhanced hydrolytic activity, three mutations were located at residues in the substrate-binding gap of subsite -3 (Arg47), subsite +1 (Ala230) and subsite +2 (Phe259). base (Fig. 1), one mutation occurs at a residue that does not interact directly with the substrate (Phe21).
[0750] Surprisingly, variants with increased hydrolysis rates were found to have significantly lower ratios...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com