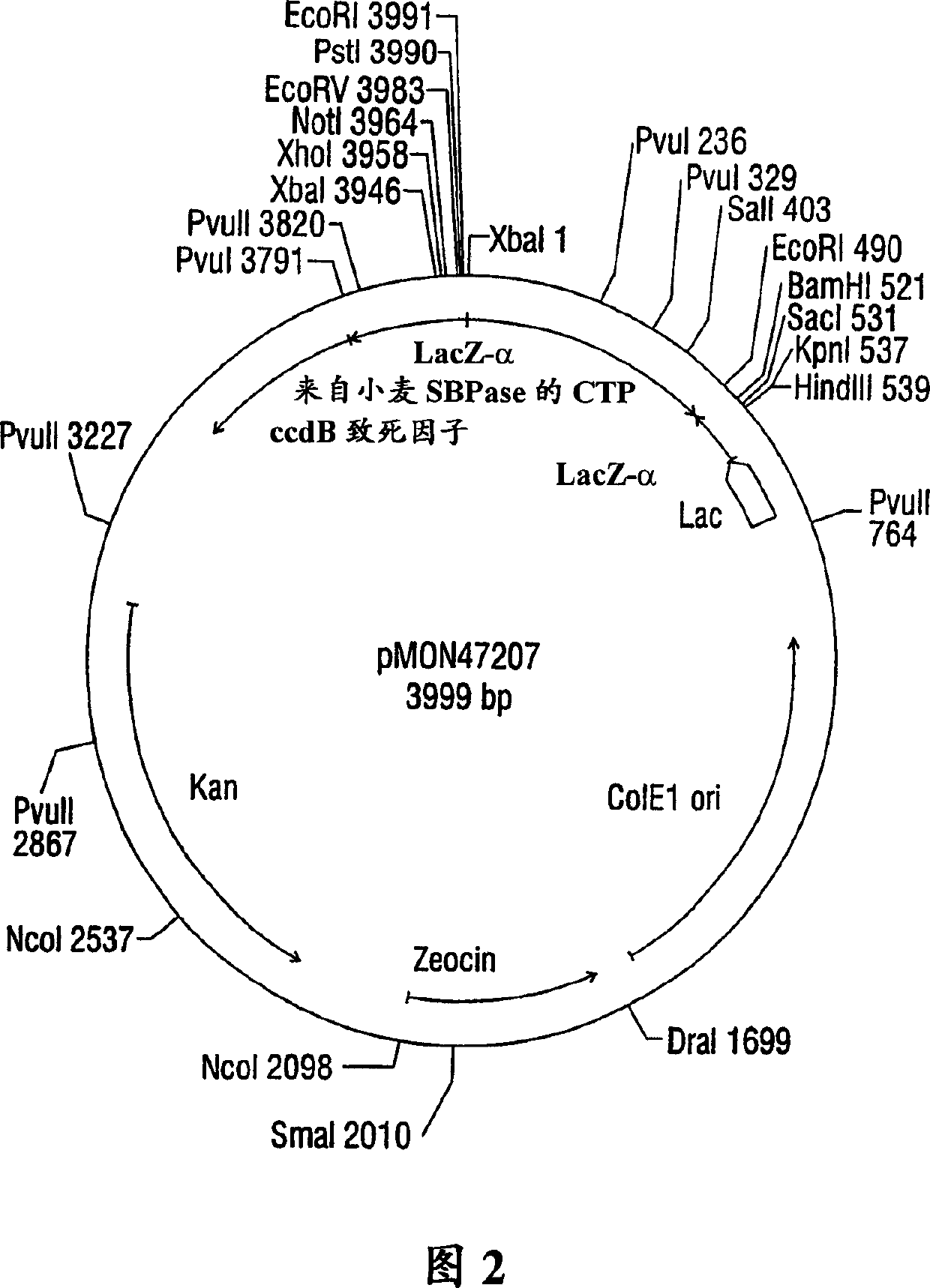
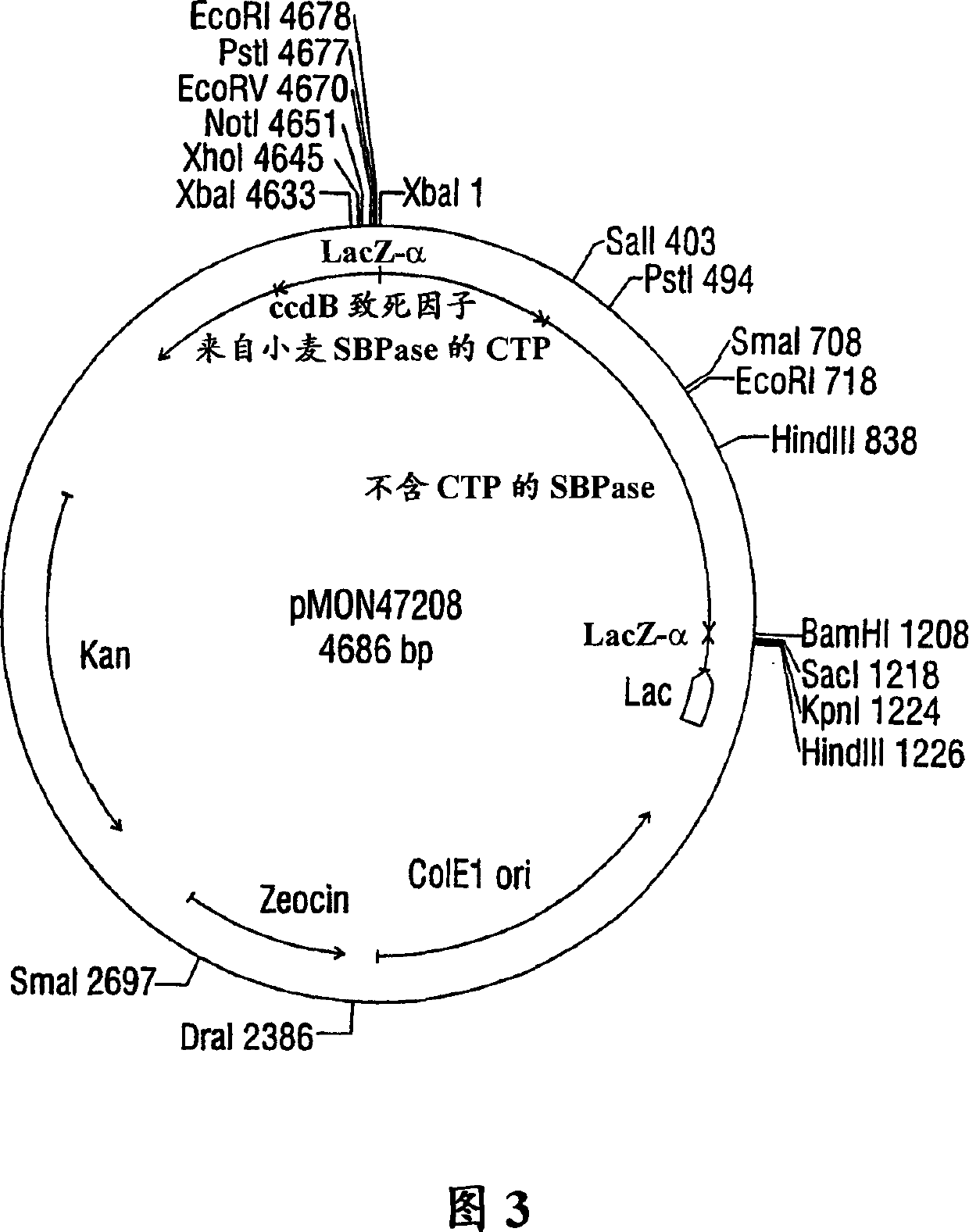
Expression of sedoheptulose 1,7-bisphosphatase in transgenic plants
A technology of sedum heptulose and diphosphatase, applied in genetic engineering, plant genetic improvement, botany equipment and methods, etc., can solve problems such as chlorosis, reduced carbon assimilation level, and reduced sugar content
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Embodiment 1
[0094] The cDNA clone of embodiment embodiment 1SBPase and excessive expression to produce antibody
[0095] To isolate the gene region encoding the mature SBPase protein (without CTP), RT-PCR reactions were performed. One microgram of common wheat (Triticum aestivum) CV OSLO leaf RNA was mixed with 100 pmol random hexamer primers (BRL / Life Technologies Inc., Gaithersburg, MD) or with 100 pmol oligo dT primers (Promega, Madison, WI), in Heat at 75°C for 5 minutes, then cool on ice. Using Superscript II TM Reverse transcriptase (BRL / Life Technologies Inc, Gaithersburg, MD), according to the manufacturer's protocol, was used for first-strand cDNA synthesis. The terminated reverse transcription reaction was diluted 1:7. Three microliters of the diluted first-strand synthesis product was mixed in 100 μL with: 100 μM of each dNTP, 50 pmol with homology to the 5' end of the gene, designed to create an NdeI cleavage site for subcloning Gene-specific primer (5...
Embodiment 3
[0099] To test the expression of wheat SBPase subunits and their assembly into active enzymes, a vector containing the following elements was constructed: the CaMV E35S promoter, the coding sequence of the complete SBPase protein including CTP, the NOS 3' termination signal, and Bacillus selection for ampicillin resistance. The SBPase gene was isolated from pMON47200 as a XbaI / BamHI fragment. The SBPase gene was ligated into XbaI / BamHI linearized pMON999 (Fig. 6) in the region with CaMV E35S, NOS 3', resulting in pMON47203 (Fig. 7). The DNA constructs were electroporated into maize protoplasts according to the method of Sheen et al. (1991). Embodiment 4 transforms the analysis of maize protoplast
Embodiment 4
[0099] To test the expression of wheat SBPase subunits and their assembly into active enzymes, a vector containing the following elements was constructed: the CaMV E35S promoter, the coding sequence of the complete SBPase protein including CTP, the NOS 3' termination signal, and Bacillus selection for ampicillin resistance. The SBPase gene was isolated from pMON47200 as a XbaI / BamHI fragment. The SBPase gene was ligated into XbaI / BamHI linearized pMON999 (Fig. 6) in the region with CaMV E35S, NOS 3', resulting in pMON47203 (Fig. 7). The DNA constructs were electroporated into maize protoplasts according to the method of Sheen et al. (1991). Embodiment 4 transforms the analysis of maize protoplast
[0100] Transform the precipitated protoplast samples with pMON47203 (SBPase) in 0.18 mL of extraction buffer (50 mM HEPES pH 7.5, 1 mM fructose diphosphate, 1 mM sedoheptulose-1,7-diphosphate, 10 mM MgCl 2 , 10mM MnCl 2 , 10mM DTT, 1% polyvinylpolypyrrolidone, 10% glycerol and C...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com