Cyclic RNAcircMDK, application thereof and liver cancer treatment preparation
A liver cancer and circular technology, applied in the direction of DNA / RNA fragments, recombinant DNA technology, medical preparations of non-active ingredients, etc., can solve the problem of no relevant research reports on functions, achieve true and reliable results, rigorous test process, and easy operation convenient effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
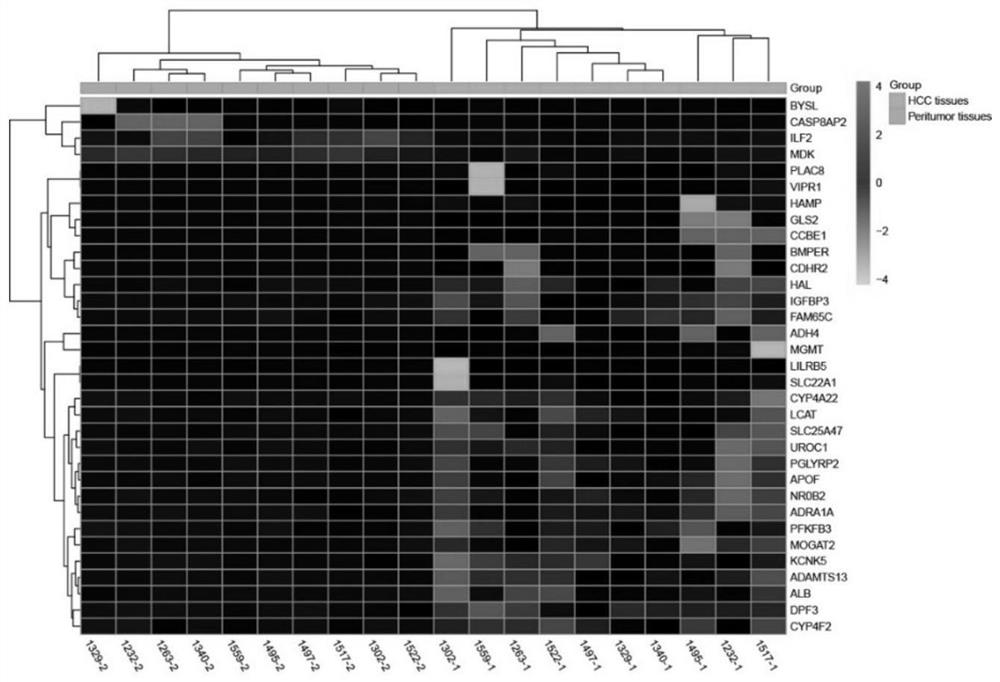
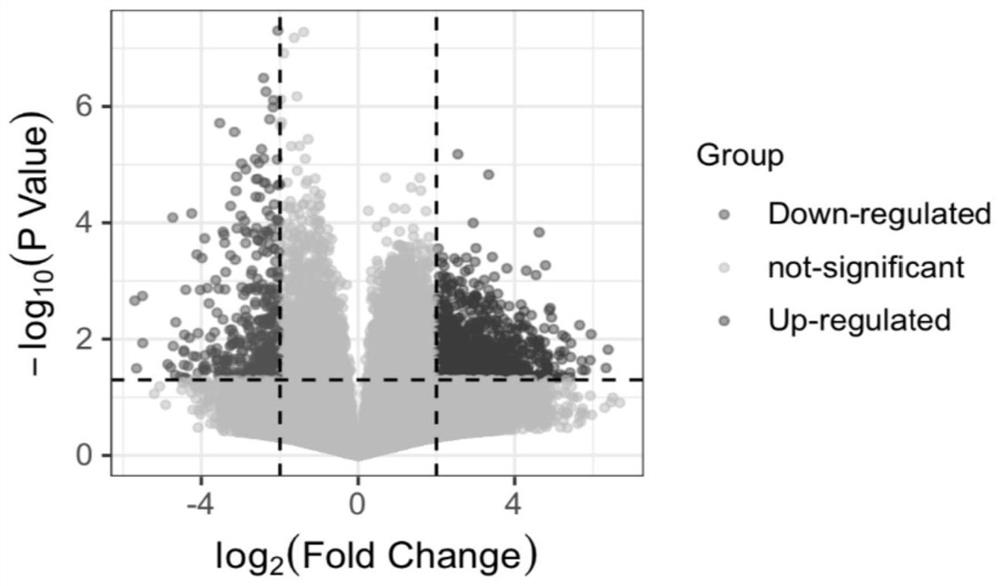
[0054] Example 1: Screening of differentially expressed circRNAs by RNA-seq experiment
[0055] RNA-Seq sequencing was performed on liver cancer tissues to screen for differentially expressed circRNAs. This part of the experiment was completed by Guangzhou Ribo Biotechnology Co., Ltd. The specific steps are as follows: after RNA extraction and quality inspection, rRNA is removed by probe, and RNaseR delinearizes RNA to construct Illumina sequencing library. Sequencing was then performed, the sequencing mode was PE150, and the sequencing data volume was 12G / 15G. For the sequencing data, first perform Raw Data filtering, de-linker sequences and low-quality reads to obtain high-quality data (Clean Data). The ribosomal RNA sequences were then removed by aligning with the ribosomal database to obtain valid reads. The effective reads are compared with the reference genome, and the comparison results are used for the identification of circular RNAs in the next step. RiboBio uses tw...
Embodiment 2
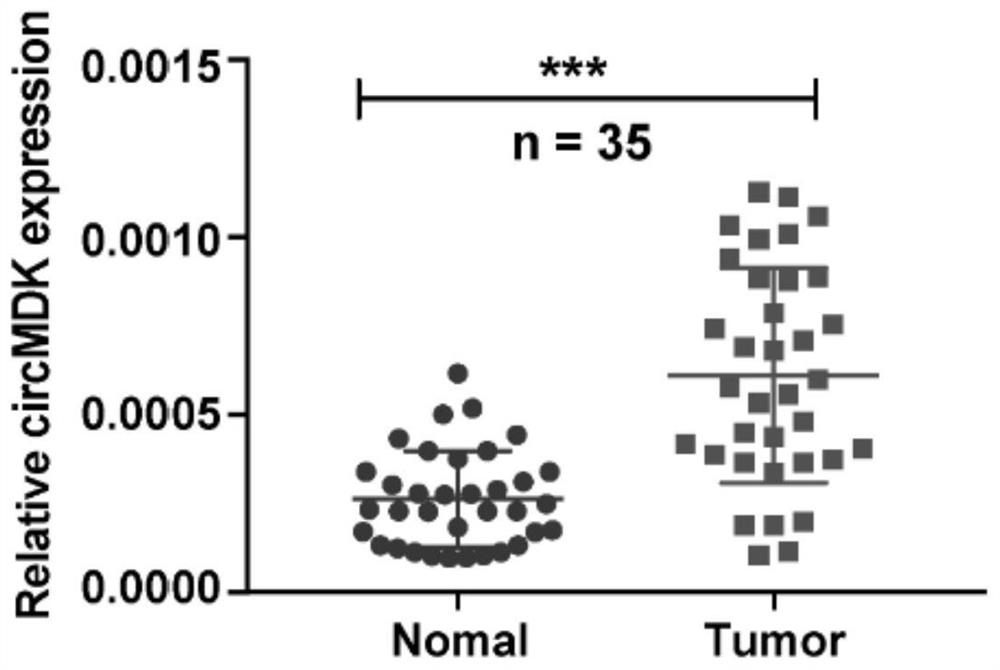
[0060] Example 2: qPCR detection of circular RNA circMDK expression in liver cancer tissue
[0061] 35 pairs of fresh liver cancer and adjacent tissues were collected to detect the expression of circular RNA circMDK by qPCR. The results are as follows image 3 shown. Circular RNA circMDK is highly expressed in cancer tissues and low in paracancerous tissues.
[0062] The expression levels of circular RNA circMDK in human normal hepatocytes LO2 and hepatoma cells HepG2, Huh7, and Hep3B were detected by qPCR. like Figure 4 As shown, the expression level of circular RNA circMDK was significantly up-regulated in Hep3B, Huh7, and HepG2 cells compared with LO2 cells.
Embodiment 3
[0063] Example 3: RNase R and Actinomycin D treatment and in situ hybridization experiments to detect the structural characteristics of circular RNA circMDK
[0064] RNase R is a highly processed 3' to 5' exoribonuclease that digests linear RNA (i.e. MDK), and resistance experiments to its digestion confirmed that circMDK has a closed loop structure ( Figure 5 ); in addition, after actinomycin D treatment, the half-life of circMDK transcripts within 24 h was more stable than that of MDK ( Image 6 ). The results of nuclear and cytoplasmic separation and RNAFISH detection together showed that circMDK was mainly localized in the cytoplasm ( Figure 7 , Figure 8 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com