Novel model for prognosis prediction and diagnosis of colorectal cancer and application thereof
A colorectal cancer and prediction model technology, applied in the field of biomedicine, to improve the evaluation and prediction ability, improve prognosis, and have broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
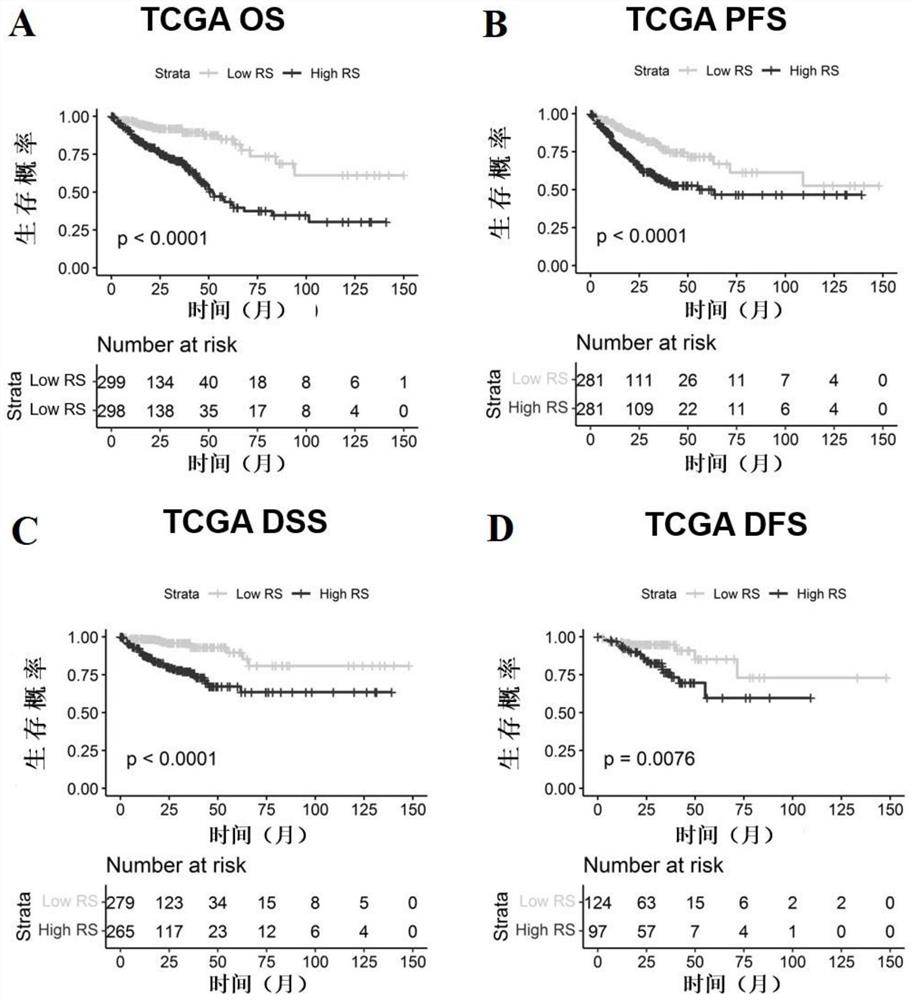
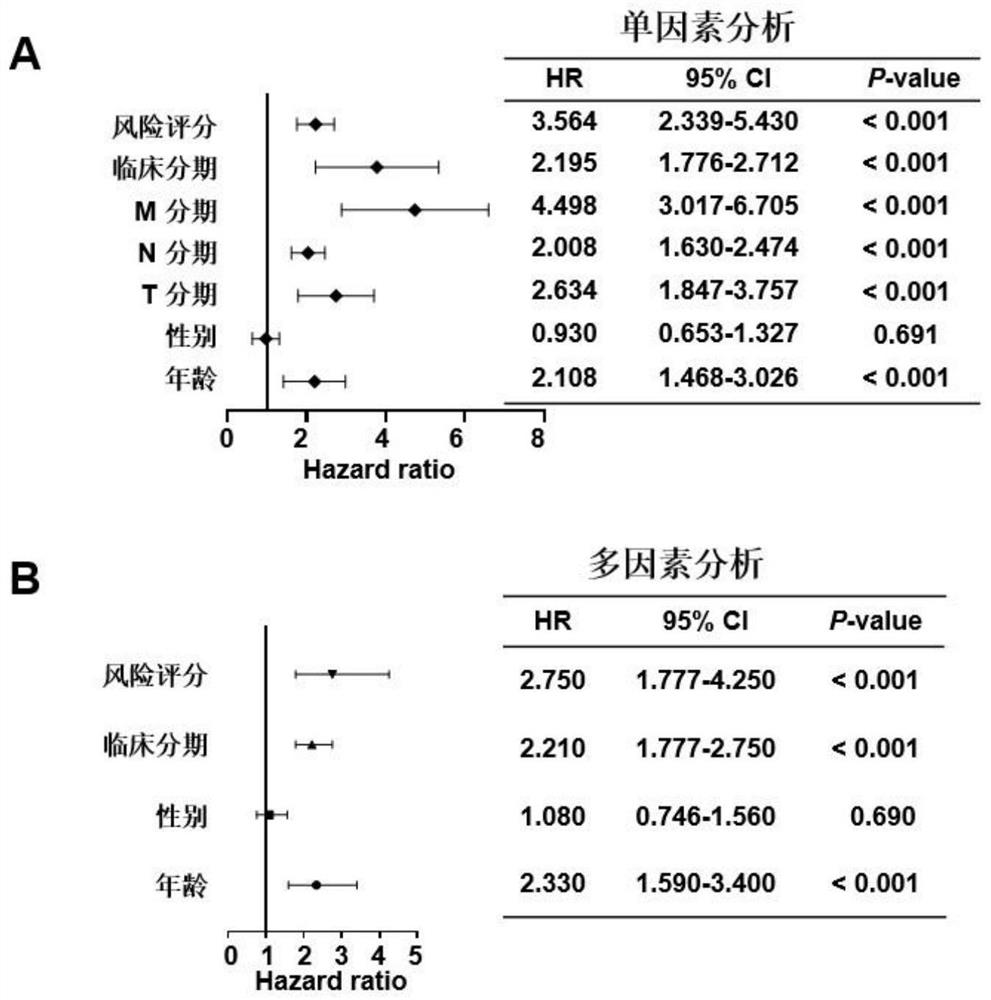
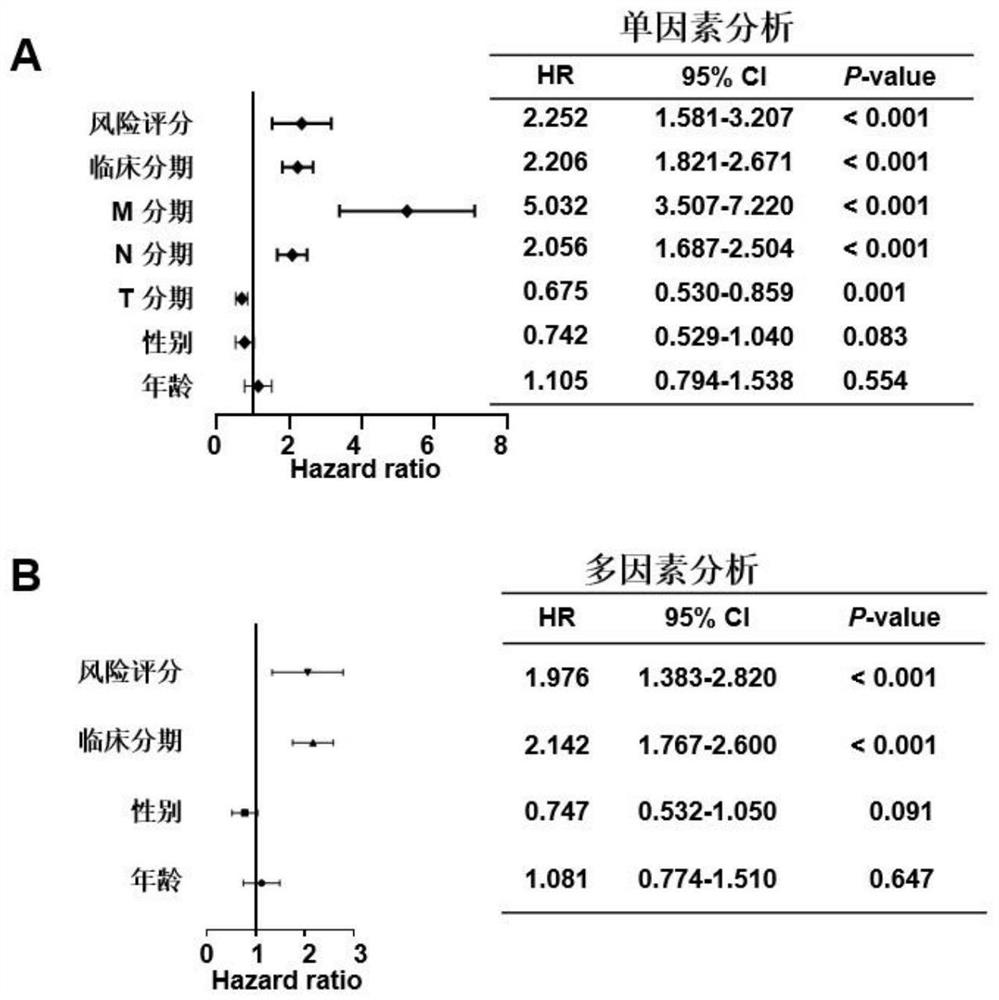
[0084] Example 1 Screening of colorectal cancer-related molecular markers, establishment and verification of an evaluation model for colorectal cancer prognosis prediction and diagnosis
[0085] 1. Data sources
[0086] The expression and clinical data of colon adenocarcinoma (COAD) and rectal adenocarcinoma (READ) in the TCGA database were downloaded from the GDC website (https: / / portal.gdc.cancer.gov / ) and merged into the colorectal cancer dataset TCGA- CRC, after removing normal, duplicate, and no follow-up information samples, a total of 597 samples were used for model building and survival analysis. In addition, two gene chip datasets, GSE39582 (n=562) and GSE37892 (n=130), were used as validation sets, and the expression matrix and clinical information were downloaded from the GEO database (https: / / www.ncbi.nlm.nih.gov / geo).
[0087] The genes involved in the cancer immune cycle were downloaded from the TIP website (http: / / biocc.hrbmu.edu.cn / TIP / ). A total of 178 char...
Embodiment 2
[0113] Example 2 Validation of the evaluation model for prognosis prediction and diagnosis of colorectal cancer established by the present invention in actual samples of colorectal cancer patients
[0114] 1. Experimental method
[0115] (1) TCGA dataset download
[0116] The expression data of normal samples in COAD and READ cohorts in the TCGA database were downloaded from the GDC website (https: / / portal.gdc.cancer.gov / ), a total of 51 cases were merged with 597 cancer samples, resulting in a total of 648 samples.
[0117] (2) Self-built clinical data set
[0118] A new clinical cohort was used in this study. The samples in this clinical cohort were from the Affiliated Hospital of Qingdao University. The inclusion criteria of patients were: colorectal cancer diagnosed by pathology and imaging, radical resection, complete clinical information, Such as age, gender, tumor size, location, TNM stage, etc., no history of other malignant tumors; the collected tissue samples were ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com