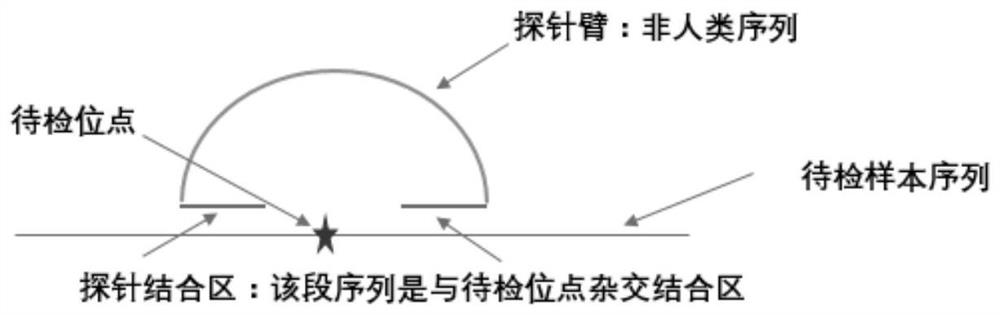
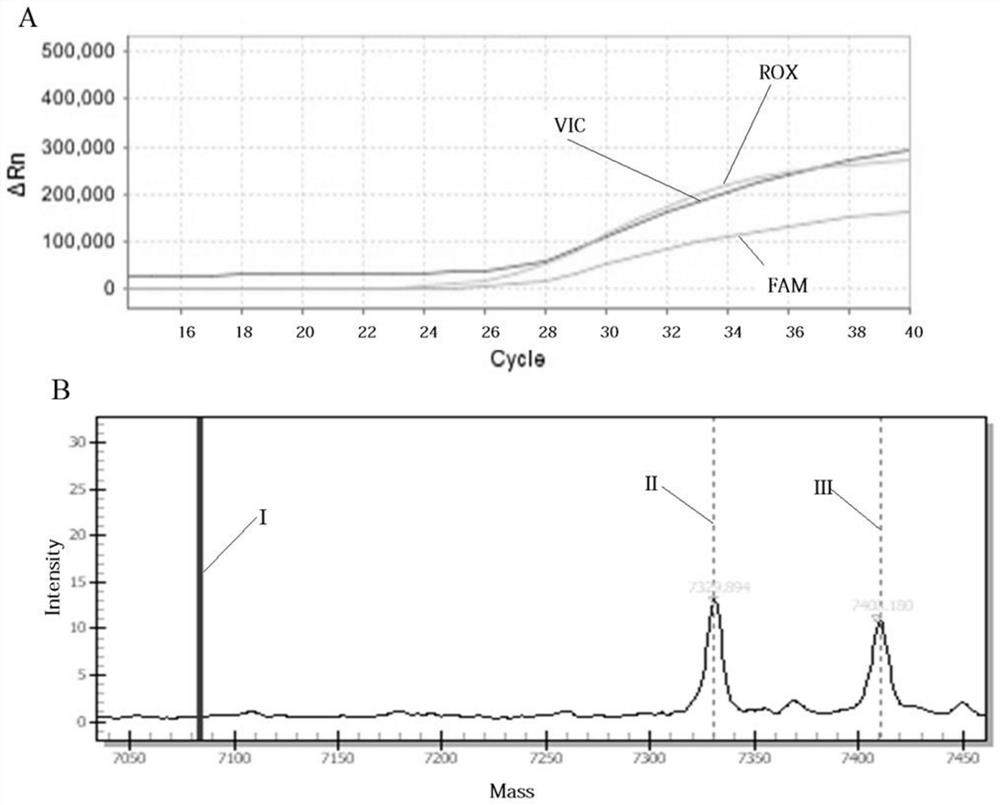
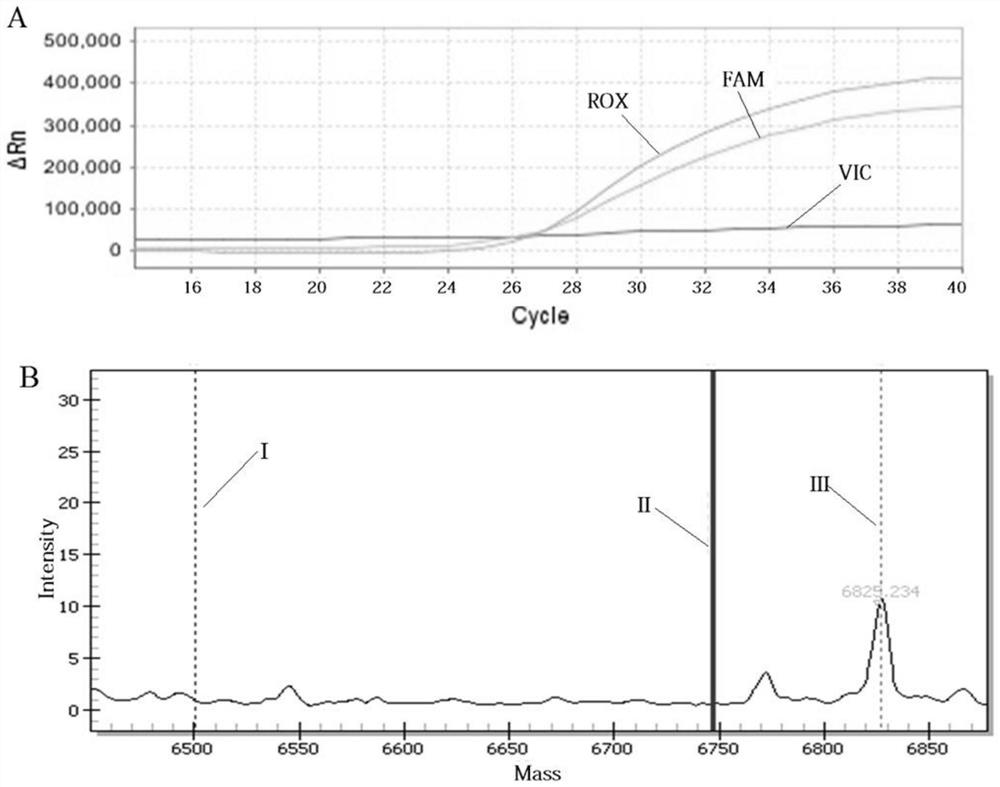
Multi-SNP locus genetic typing method based on nMALDI-TOF technology
A genotyping method and locus technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve problems such as the inability to produce test results on the same day, and overcome the problem of non-specific binding of primers, multi-gene loci The effect of shortening detection and experiment time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1: Multi-SNP locus genotyping method based on nMALDI-TOF technology
[0033] 1. Hybridization reaction
[0034] 1.1 Take out the ice box, sample DNA to be tested, probe MIX, and hybridization buffer, thaw at room temperature, vortex and mix well after thawing, centrifuge briefly, and place on ice box for use. Prepare and label the corresponding number of PCR tubes.
[0035] 1.2 Prepare a hybridization reaction system (10ul) in a 200ul PCR tube according to the following table:
[0036] Table 1 Hybridization reaction system
[0037]
[0038] Note: When the sample is peripheral blood, "X" indicates the sample volume of 10-50ng DNA, and 50ng is recommended.
[0039] 1.3 Mix by vortexing or pipetting, centrifuge briefly, place it on the PCR machine, and run the following program:
[0040] Table 2 Hybridization reaction program
[0041]
[0042] Note: The running time of the instrument is about 95min.
[0043] 2, extension, connection
[0044] 2.1 Take o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com