Linkers for characterizing polynucleotides and uses thereof
A technology of polynucleotides and adapters, which is applied in the determination/inspection of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problems of sequencing time length, sequencing speed reduction, and affecting the output of sequencing data, etc., to achieve The effect of avoiding ATP empty consumption and improving sequencing efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
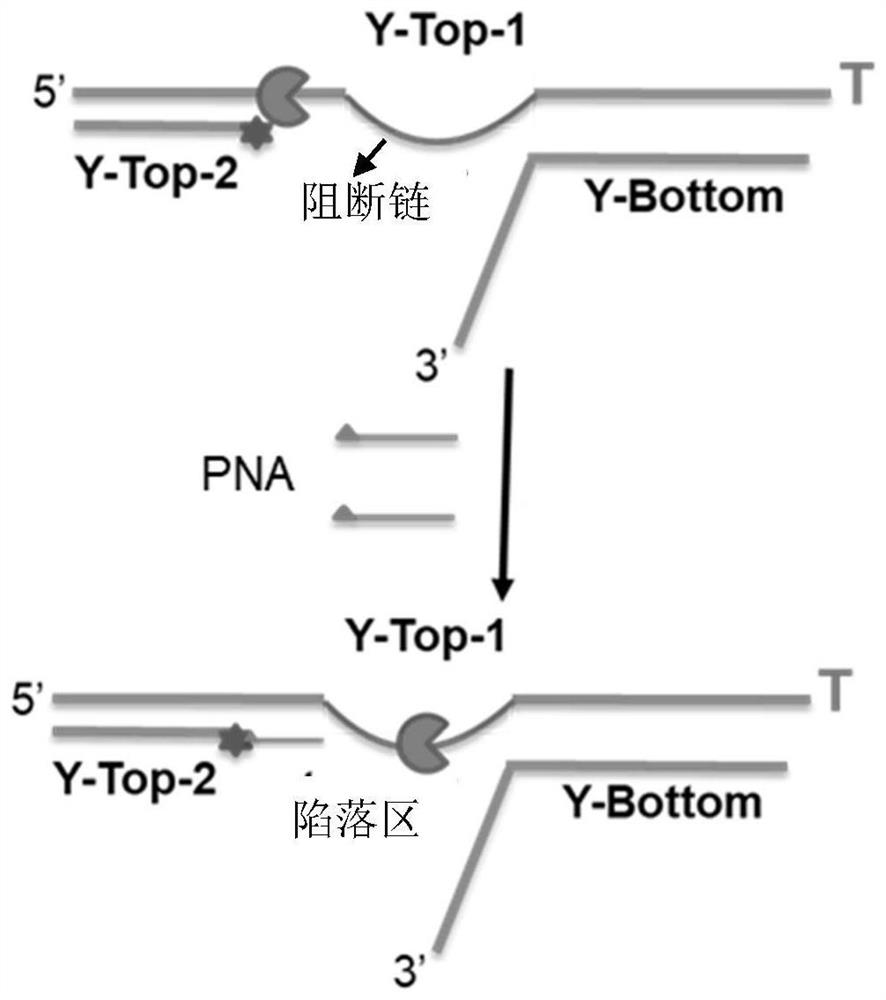
[0083] The invention provides a preparation method of the complex, comprising:
[0084] S1: binding the Y1 chain containing L'-S to the motor protein, and the binding region is located in the L' chain;
[0085] S2: adding the PNA-R chain comprising the complementary chain L" to obtain the complex, wherein the motor protein is driven to the blocking chain by the PNA-R chain;
[0086] Preferably, the method includes
[0087] S101: make contain D 1 '-L'-S-D 2 'Y1 chain, containing D 1 ” of the Y2 chain and contain D 2 The annealed product of the YB chain of " is combined with the motor protein, and the binding region is located at the L' chain of the annealed product;
[0088] S102: Adding the PNA-R chain containing the complementary chain L" to obtain the complex, wherein the motor protein is driven to the blocking chain by the PNA-R chain.
[0089] polynucleotide
[0090] Polynucleotides such as nucleic acids are macromolecules containing two or more nucleotides. A po...
Embodiment 1
[0152] Example 1: Preparation of a Y adapter-enzyme complex that reduces ATP empty consumption
[0153] SEQ ID NO: 1GCGGAGTCAAACGGTAGAAGTCG
[0154] SEQ ID NO:2TAACGTATTC
[0155] SEQ ID NO:3ACTGCTCATTCGGTCCTGCTGACT
[0156] SEQ ID NO:4CGACTTCTACCGTTTGACTCCGC
[0157] SEQ ID NO:5GTCAGCAGGACCGAATGA
[0158] SEQ ID NO:6GAATACGTTAGCGG, wherein SEQ ID NO:6 consists of PNA
[0159] SEQ ID NO:7GCAGTAGTCCAGCACCGACC
[0160] SEQ ID NO:8
[0161] GTFDDLTEGQKNAFNIVMKAIKEKKHHVTINGPAGTGKTTLTKFIIEALISTGETGIILAAPTHAAKKILSKLSGKEASTIHSILKINPVTYECNVLFEQKEVPDLAKARVLICDEVSMYDRKLFKILLSTIPPWATIIGIGDNKQIRPVDPGENTAYISPFFTHKDFYQCELTEVKRSNAPIIDVATDVRNGKWIYDKVVDGHGVRGFTGDTALRDFMVNYFSIVKSLDDLFENRVMAFTNKSVDKLNSIIRKKIFETDKDFIVGEIIVMQEPLFKTYKIDGKPVSEIIFNNGQLVRIIEAEYTSTFVKARGVPGEYLIRHWDLTVETYGDDEYYREKIKIISSDEELYKFNLFLGKTCETYKNWNKGGKAPWSDFWDAKSQFSKVKALPASTFHKAQGMSVDRAFIYTPCIHYADVELAQQLLYVGVTRGRYDVFYV
[0162] The complex is formed by the hybridization of 4 different strands;
[0163] The first stran...
Embodiment 2
[0176] Embodiment 2: ATPase activity detection
[0177] Firstly, the NADH reaction mixture was prepared, and the reaction mixture was mainly prepared according to Table 1 below. After the preparation was completed, the reaction mixture was turned horizontally at room temperature and incubated for 10 minutes.
[0178] Table 1 Preparation of NADH reaction mixture
[0179]
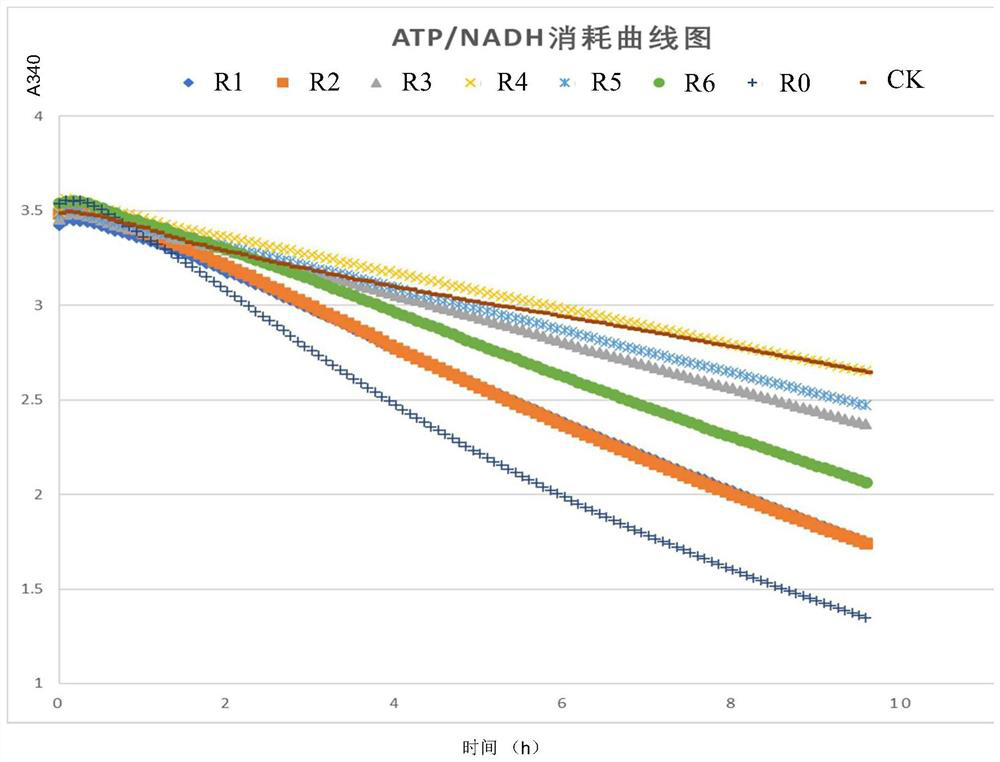
[0180] After that, add 112.5 μL NADH reaction mixture in 96-well plate, 37.5 μL (20nM) Y adapter-enzyme complex thing (which is Any one of sample 1 and sample 2 after purification in embodiment 1 ), and then put it into a UV-visible spectrophotometer to measure the absorbance value at 380nm, and set the temperature to 34°C; detect 200 cycles, each cycle is 5 minutes. The collected data were drawn into a standard curve and the ATP consumption value was obtained through the slope of the standard curve.
[0181] The result is as image 3 and Table 2 (adding complex 10h). No addition of PNA-R was used...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com