Metagenome data analysis method
A metagenomics and analysis method technology, applied in sequence analysis, instrumentation, biostatistics, etc., can solve the problem of lightening, and achieve the effect of reducing pressure, improving data utilization, and reducing sequencing costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0089] Embodiment 1 uses the method analysis data of the present invention
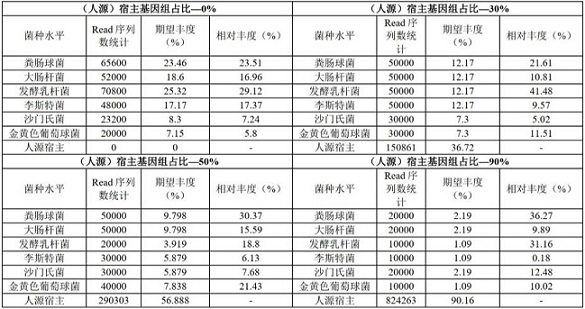
[0090] 1. Four sets of metagenomic standards containing 0%, 30%, 50% and 90% of the human host ratio, each containing Enterococcus faecalis, Escherichia coli, Lactobacillus fermentum, Listeria monocytogenes, Salmonella enterica, Staphylococcus aureus, and the expected abundance of each bacteria in each standard is known, through the experimental library Preparation, using the third-generation nanopore sequencer model QNome-9604 for sequencing to obtain the original long-read sequencing data, the maximum read length of the data reaches 16Kb, and the average length is 7.8kb; use Porechop software and NanoFilt software to remove experimental constructs Linkers and barcode sequences added in the library process, filter low-quality read sequences below Q5 and length less than 100bp;
[0091] 2. For each read sequence in the data set of the sequence obtained in step 1, perform 20 sliding extractions (N=2...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com