Three-generation library building sequencing method for whole genome full-length amplification based on novel coronavirus
A whole-genome and virus technology, applied in the field of third-generation library construction and sequencing, can solve the problems of not fully exploiting the advantages of long-read lengths, and not studying long-read length sequences, so as to reduce waste, prevent amplification bias, and improve experimental efficiency. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] 1. Primer design and synthesis
[0045] Searching for the new coronavirus on NCBI, a total of 74 new coronavirus sequences were downloaded and obtained. By checking the sequence integrity and deleting sequences with a length of less than 20k, a total of 43 complete new coronavirus sequences were obtained. All 43 complete genome sequences were compared to obtain a consensus sequence, and the consensus sequence of the new coronavirus genome is shown in SEQ ID NO: 1.
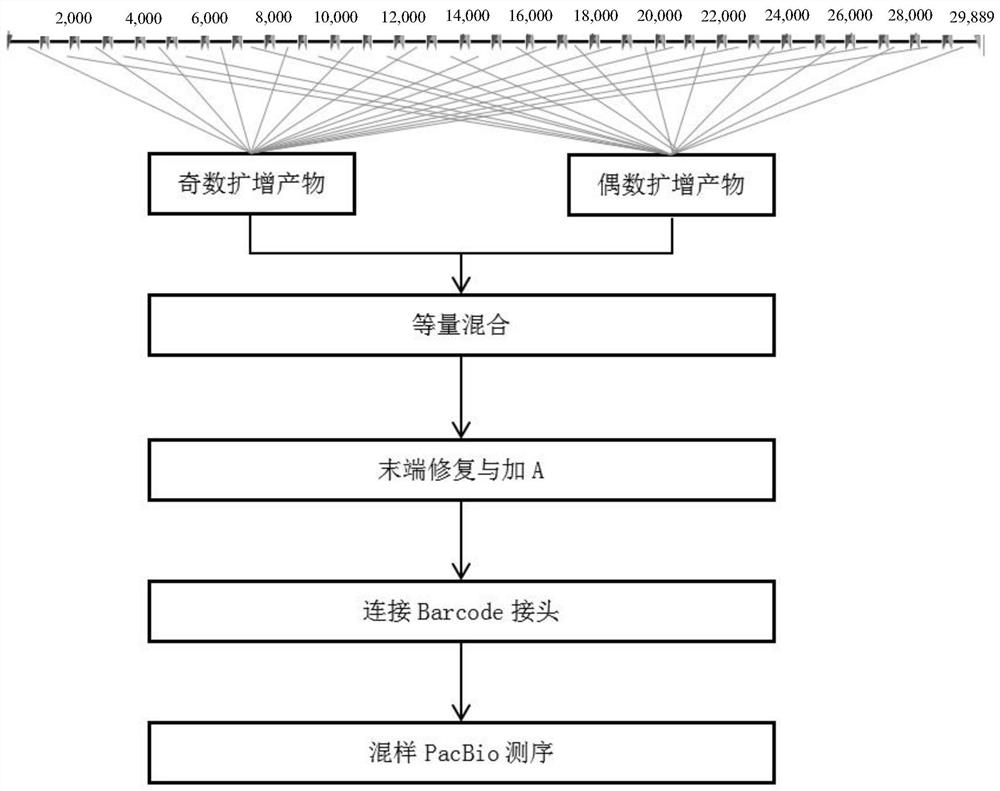
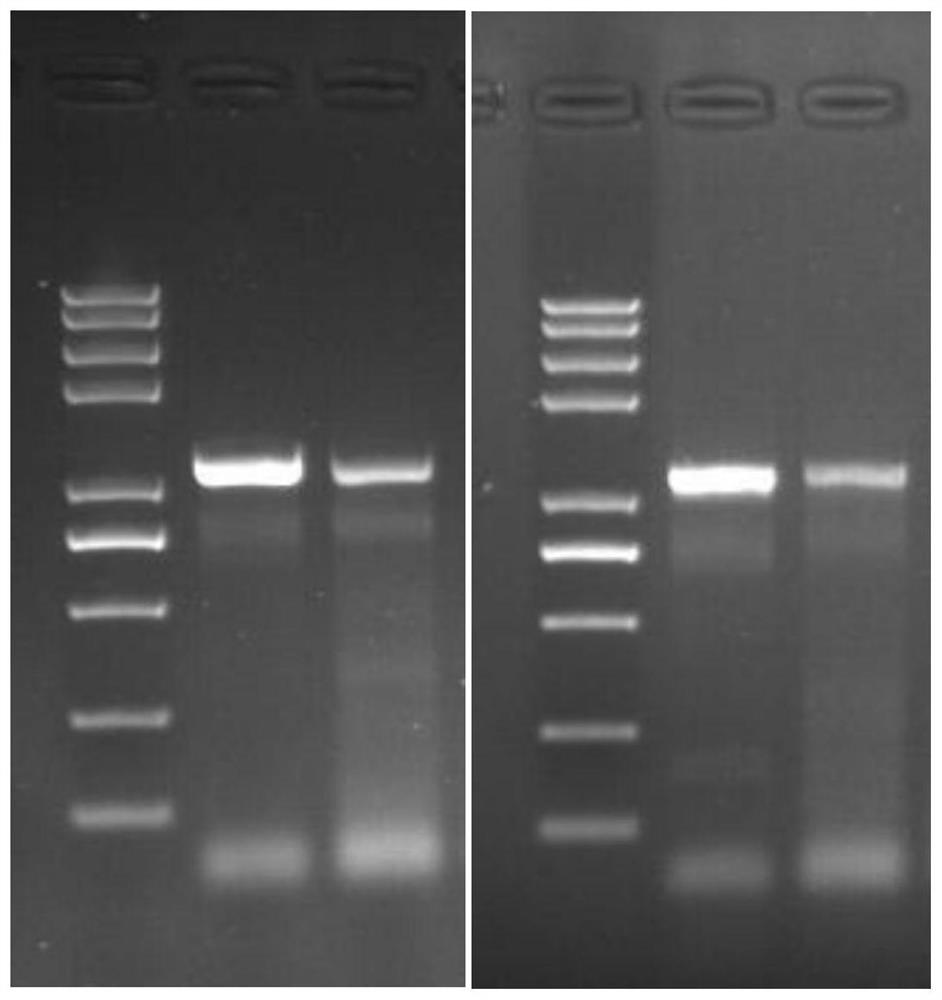
[0046] Based on the consensus sequence, a total of 30 pairs of primers were designed. The primers covered the entire 2019-nCoV genome. The primer positions were designed based on the conserved regions of the genome. There was an overlap of about 100 bp between each pair of primers. The size of the PCR product amplified by each pair of primers was 0.9-1.2 k. Primer design positions such asfigure 1 As shown, the primers in this scheme include a total of 30 pairs, and the amplified fragment size of each pair o...
Embodiment 2
[0081] Example 2: Comparison of virus data ratios in the amplified sequencing data
[0082] Table 6 shows the proportion of virus data in the sequencing data before and after the amplification of nucleic acid samples WD1 and WD2 extracted from oral swabs in Example 1.
[0083] Table 6:
[0084] sample name Unamplified Data Virus Ratio Amplified Data Virus Ratio Increase multiple WD1 0.7622% 71.20% 93.4 WD2 0.0014% 13.79% 9850
[0085] It can be seen that, after the sample is amplified by improving the amplification method provided by the present invention, the data virus ratio is increased to hundreds of times to hundreds of thousands of times, and the virus information can be enriched, which greatly reduces the sequencing data. waste.
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com