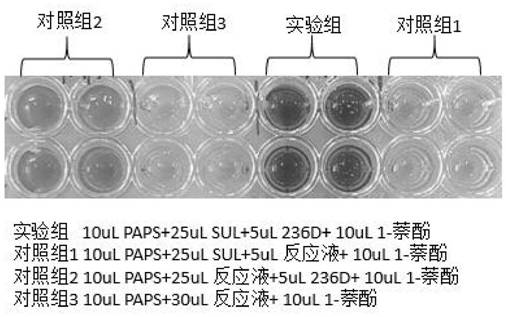
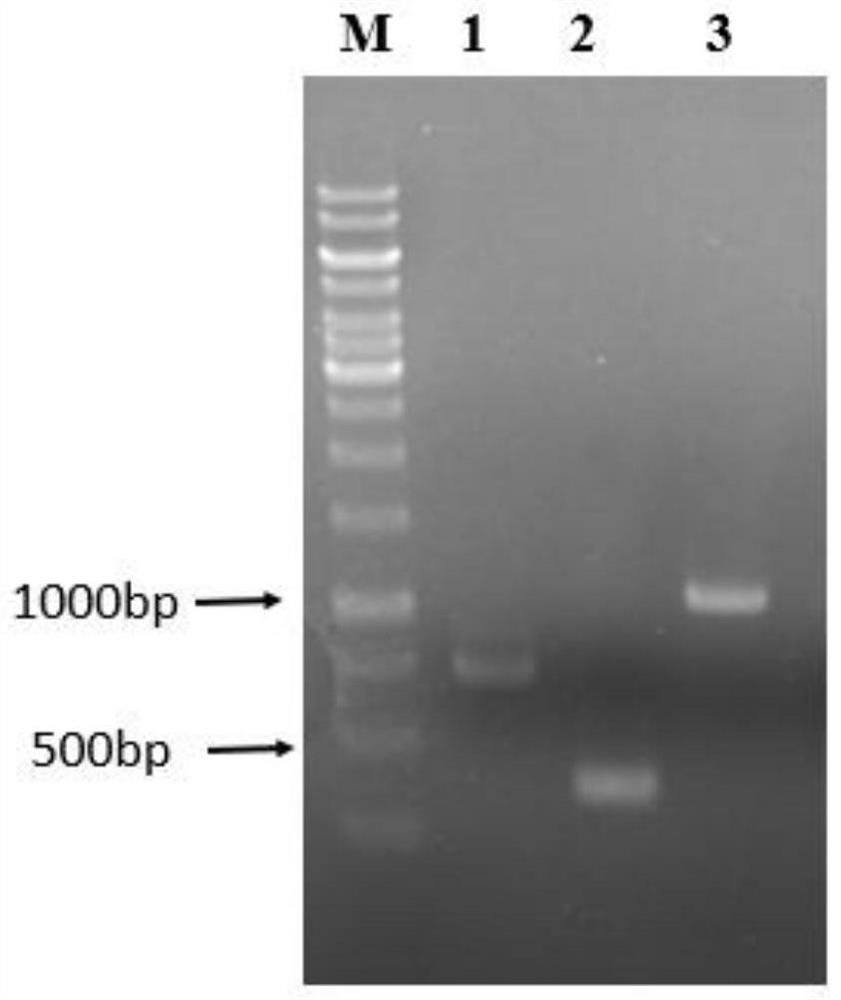
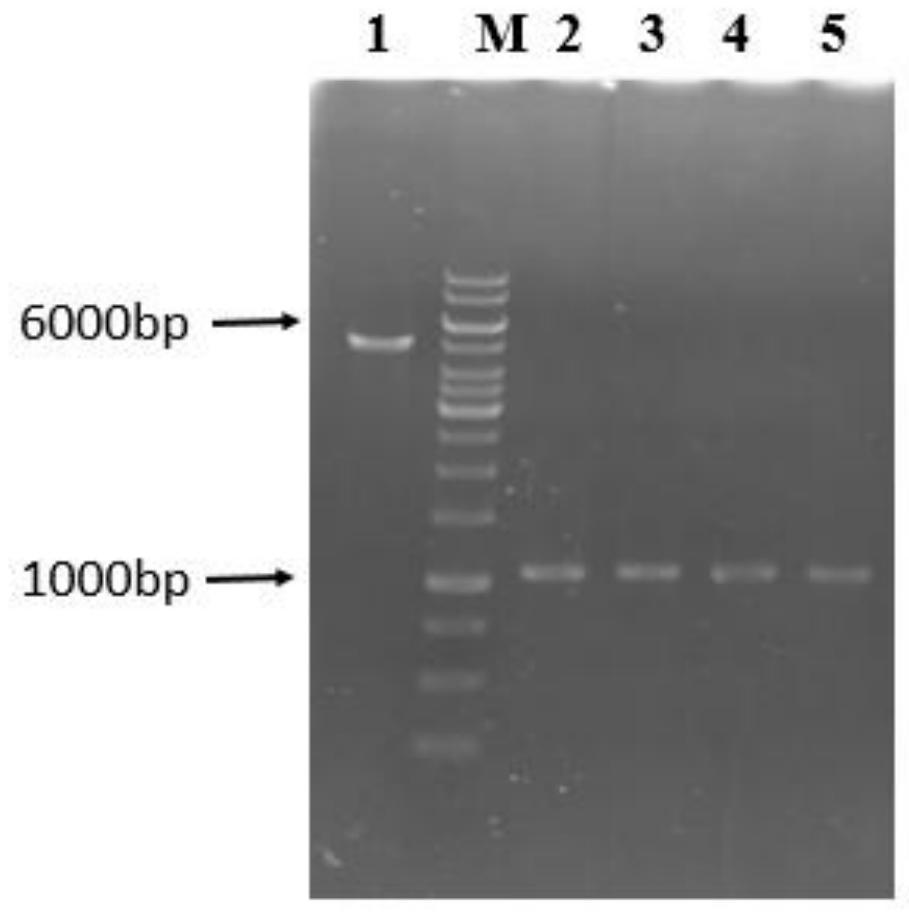
High-specificity coupling enzyme YND mutant of PAP based on molecular docking, method and application
A molecular docking, high-specificity technology, applied in the biological field, can solve the problems of large amount of screening experiments, high price, single buffer, etc., and achieve the effect of reducing screening workload, improving substrate specificity, and reducing detection costs.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0037] The embodiments of the present invention will be described in detail below. It should be noted that the embodiments are illustrative, not restrictive, and cannot limit the protection scope of the present invention.
[0038] The raw materials used in the present invention, unless otherwise specified, are conventional commercially available products; the methods used in the present invention, unless otherwise specified, are conventional methods in the art.
[0039] A highly specific coupling enzyme YND mutant of 3',5'-adenosine diphosphate based on molecular docking, the gene sequence of the YND mutant is SEQ ID NO.2, SEQ ID NO.3 or SEQ ID NO.3 ID NO.4, the amino acid sequence of the YND mutant is SEQ ID NO.6, SEQ ID NO.7 or SEQ ID NO.8.
[0040] Preferably, the molecular docking software Vina 1 It is used to design YND with high specificity for PAP substrates, and the three-dimensional structure of YND uses PDB ID 1K9Y 2 The protein structure of YND; the mutant of YND ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com