Primer group, kit and method for detecting cattle horn-free gene
A technology of primer sets and kits, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/inspection, etc., can solve problems such as inaccurate typing, time-consuming, and inability to achieve rapid detection, etc., and achieve improvement The effect of the hornless gene frequency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
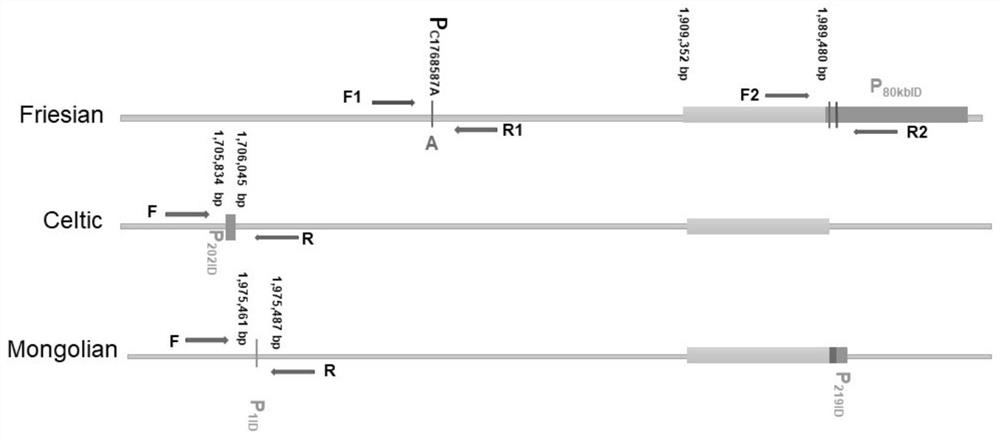
[0090] Example 1 The design and optimization process of the primer group
[0091] According to the three kinds of unopened gene sequences, the layout primer groups I, II, III are designed. Among them, the primer group I is based on P 202ID The specific sequence of the 2 alleles of the site is optimized, using the differentiality of the repeating sequence; the primer group II is based on P 219ID The specific sequence of the 2 alleles of the site is optimized, and 6 bases are replaced by 7 bases at upstream 628 bp (resulting in 1 bp length difference); primer III is based on P 80kbID The specific sequence of the 2 alleles of the site is optimized, and 2bp is deleted in the repeating sequence.
[0092] The optimized design process of primer III is specifically described for example. Specifically, the inventors have the Sanger sequencing results of the Friesian unopened gene P80kbid site, and the original 80KB sequence represents the original 80KB sequence, square bracket "[]" represe...
Embodiment 2
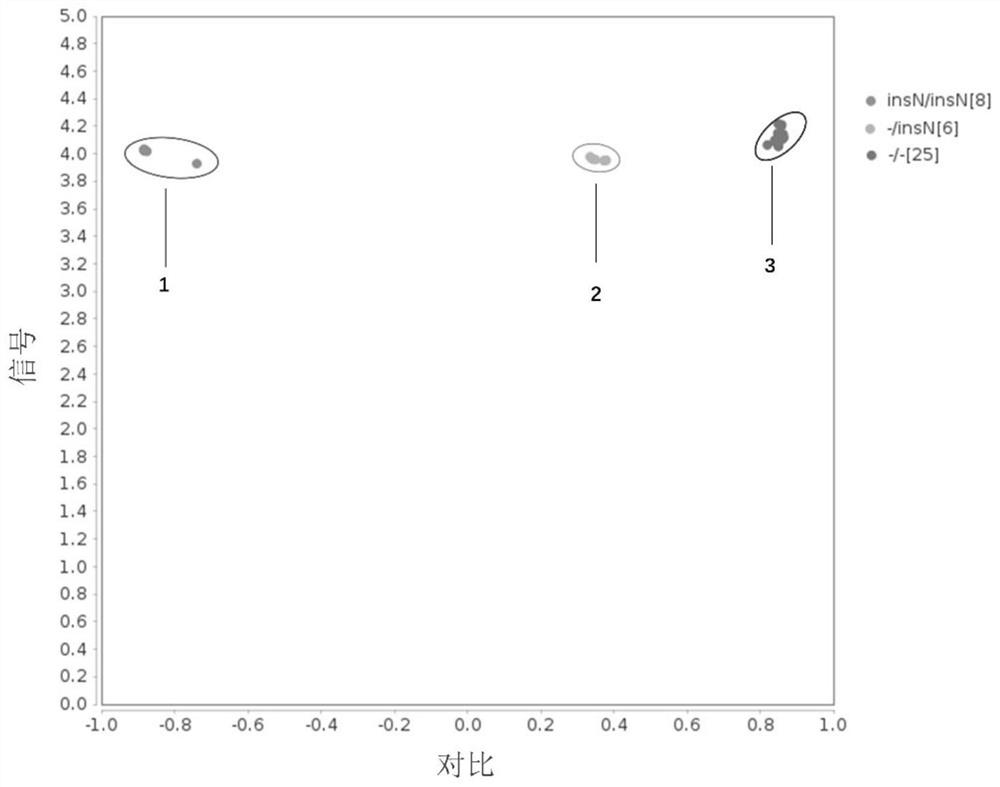
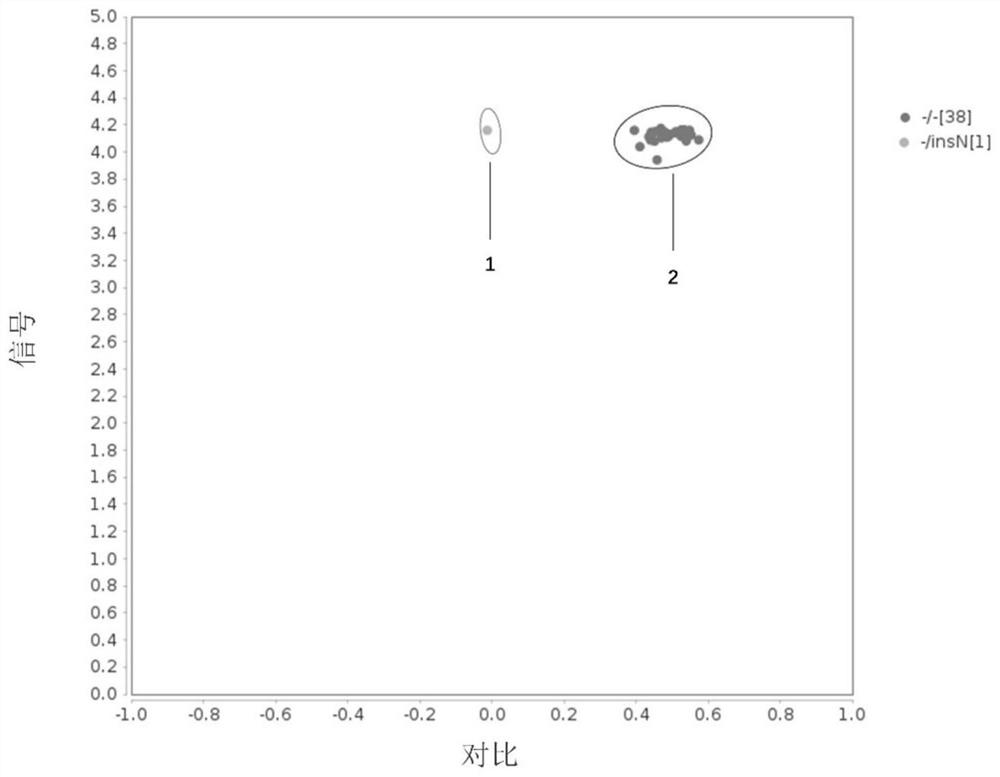
[0099] Example 2 Terminal detection of three kinds of cattle angle genes
[0100] Collect 39 bulls freeze semen of different varieties, and extract genomic DNA by high salt method. Among them, the samples include 9 cows, varieties are Holinstein cattle and Juan Hao cow; bull cows 17, varieties include Angers Niu, Flewiki Niu, Charlottle Niu, Limuzi and Simmental Bull; There are 13 three rivers, Qinchuan cattle and Mongolia.
[0101] SNP is performed based on the principle of KASP competitive allele specific PCR, wherein the PCR reaction is completed on the microfluidic chip (Beijing Bo Otry Biotechnology Co., Ltd., the product number G020010), the method steps are:
[0102] 1) On a dedicated microfluidic chip, genomic DNA (5 ~ 10 ng) to be detected in the chip well is injected;
[0103] 2) For 4 sites, the PCR amplification reaction mixture is separately configured. In the reaction mixture of each site, a site-specific primer (Table 1), PCR premix, and double steamed. The configur...
Embodiment 3
[0115] Example 3 General detection verification KASP method accuracy
[0116] In order to verify the accuracy of KASP detection results, all 39-head sample PCR amplification, and detect the angular genotype by agarose gel electrophoresis or Sanger sequencing.
[0117] (1) PCR primer
[0118] In response to different unopened gene sites, the primers designed in the literature and Primer3.0 (http: / / bioinfo.ut.ee / primer3 / ) are used to detect, and the primer details are shown in Table 3.
[0119] Table 3 primer sequence and amplification fragment length
[0120]
[0121] (1) PCR amplification
[0122] PCR reaction system: Total volume 25 μL, of which 10 × buffer 2.5 μL, DNTP mixture (2.5 mM) 2 μL, TAQ enzyme (5 U / μL) 0.5 μL, 5 μm of the upper and lower proders (20 μm), 1 μL of genomic DNA template (about 50 ng), with double steamed water to make up 25 μL.
[0123] PCR reaction conditions: 5 min at 94 ° C; then 35 cycles, 94 ° C denaturation 30SEC, and the annealing temperature of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com