A kind of dehydrogenase mutant l283v/l286v and its preparation method and application
A L286V, mutant technology, applied in the field of biomedicine, can solve the problems of undiscovered enzyme design and transformation, and achieve the effects of increasing substrate diversity, reducing reactant components, and improving dehydrogenase activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] The establishment of the tertiary structure model of MesPDH enzyme:
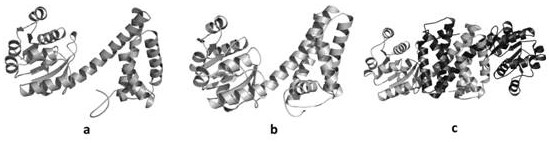
[0038] 1. The analysis was derived from Mesorhizobia ( Mesorhizobium sp. L48C026A00)’s 6-phosphogluconate dehydrogenase MesPDH, using the homology modeling tool MODELLER to carry out homology modeling on MesPDH, obtain the monomer MesPDH enzyme model Ma, evaluate the model, and evaluate all amino acids in the reasonable Lagrangian conformation diagram region, the proportion of amino acids falling in the optimal region is as high as 97.3%, the model is reasonable, and the model Ma is as follows figure 1 as shown in a.
[0039] 2. Use the online homology modeling tool Swiss-model to carry out homology modeling on MesPDH, obtain the dimer MesPDH enzyme model Mb, evaluate the model, and evaluate all amino acids in the Lagrange conformation diagram. The proportion of amino acids in the region is as high as 94.6%, the model is reasonable, and the model Mb is as follows figure 1 as shown in b.
[0040] 3...
Embodiment 2
[0042] Molecular docking simulation predicts the binding conformation of each substrate and enzyme and selects the site:
[0043] 1. Molecular docking simulation
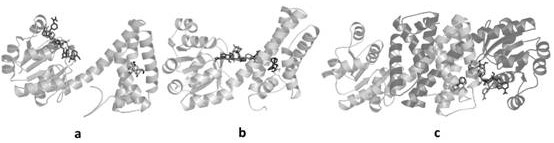
[0044] The molecular simulation software Discovery Studio was used to locate the active pocket centers of the three modeling models, and the molecular docking software AutodockVina was used to dock the MesPDH enzyme with Mesmin, glucose, gluconic acid, NAD+, and NADH, and the protein conformation analysis tool PyMOL , for each docking complex analyzed.
[0045] 2. Analysis of docking results
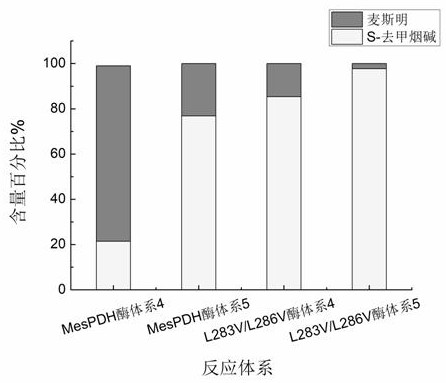
[0046] Such as figure 2 As shown, the MesPDH enzyme model Ma, Mb and Mc were docked with the four reactant molecules respectively, and it was found that the positions of myosmine, glucose and gluconic acid all fell in similar positions, and NAD+ and NADH were located in another pocket position , from the model Mb, it is speculated that the MesPDH enzyme is most likely to exert its enzyme activity in the form of a dimer, wh...
Embodiment 3
[0050] Obtaining of recombinant wild enzyme strain BL21(DE3) / pET-32a-MesPDH:
[0051] The whole gene was synthesized and optimized by Escherichia coli codon preference to obtain the MesPDH enzyme gene, its nucleotide sequence is as shown in SEQ ID NO: 4, which was connected to the pET-32a plasmid, and the obtained recombinant plasmid was named pET-32a-MesPDH , the plasmid was transformed into Escherichia coli BL21 (DE3), and the recombinant strain was named BL21 (DE3) / pET-32a-MesPDH. The amino acid sequence of the wild type MesPDH enzyme expressed by the recombinant wild enzyme strain is shown in SEQ ID NO:3.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com