Method for screening mutation of red transcription factor EKLF gene and application thereof
A gene and detection method technology, which is applied in the field of screening erythroid transcription factor EKLF gene mutation, can solve the problems of cumbersome operation steps, high detection cost, and long time consumption, and achieve the effect of simple operation, high sensitivity, and fast speed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
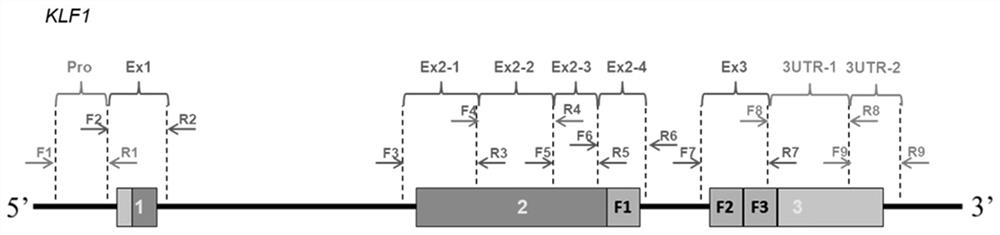
[0042] According to the EKLF gene sequence in GenBank, the present invention uses Primer Express3.0 software to design 9 pairs of specific primers covering the non-coding region of EKLF gene (promoter region, splicing site, 5'UTR and 3'UTR) And all coding regions (exon 1, exon 2 and exon 3), the shortest amplicon fragment length is 179bp, the longest is 348bp, and its specificity was initially identified by NCBI's BLAST function. The positions of the primers cover each other, avoiding screening "dead spots".
[0043] The 9 pairs of upstream and downstream primers are as follows:
[0044] 1) The primers for the upstream and downstream of the EKLF gene promoter region are: 5'-TACCCAGCACCTGGACCCTC-3', 5'-GAGGCTGTGATAGCCCCTTCG-3';
[0045] 2) The primers for the upstream and downstream of the exon 1 region of the EKLF gene are: 5'-CTAAGGACAGAGAGGAGCCC-3', 5'-CAGCCAGCCCACCTAGAC-3';
[0046] 3) The primers for the upstream and downstream regions of exon 2-1 of the EKLF gene are: 5...
Embodiment 2
[0054] The present invention discloses a reaction system and PCR reaction conditions containing the above-mentioned PCR primer set, wherein the reaction system mainly includes the following components: 9 sets of primer pair mixture (10 μM) described in Example 1, negative quality control substance, 10×Taq buffer (containing 20mM Mg2+), 2.5nM dNTPs, 5U / μl Taq DNA polymerase, 1×LC Green, template DNA, 5M Betain, DMSO and deionized water.
[0055] Wherein the reaction condition is:
[0056] 1) PCR amplification conditions: pre-denaturation at 95°C for 5 minutes; 45 cycles (melting: 95°C for 30 seconds, annealing: 63-68°C for 30 seconds, extension: 72°C for 30 seconds); denaturation: 95°C for 1 minute, Keep warm at 4°C.
[0057] 2) Reaction conditions for HRM analysis with a resolution melting curve analyzer (LightScanner HRⅠ96): the initial temperature "Start Temp" is 55°C, the end temperature "End Temp" is 98°C and the standby temperature "Hold Temp" is 42°C, When the temperat...
Embodiment 3
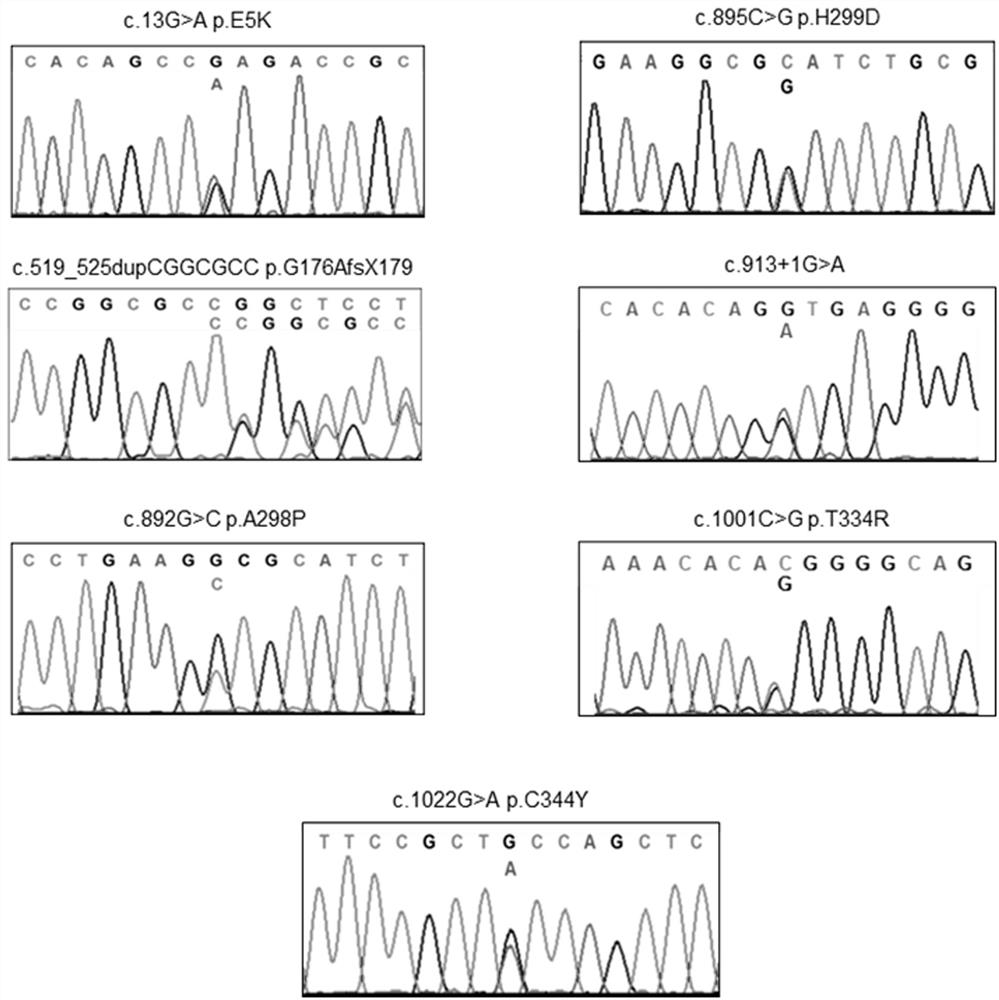
[0058] Example 3 Taking the mutation screening of EKLF gene promoter region as an example, the HRM detection of EKLF gene mutation was carried out.
[0059] 1) Specifically include the following:
[0060] Step 1, DNA preparation of the sample to be tested: collect the peripheral blood sample of the subject to be tested, and extract the sample DNA by column method;
[0061] Step 2, Aliquot the reaction solution: mix the upstream and downstream primers of the EKLF gene promoter region and the PCR reaction solution, and divide it into a 96-well PCR plate of Bio-Rad, each well includes 1.0μL of 10×Taq buffer (containing 20mM Mg2+) , 2.5nM dNTPs 0.4μL, 5U / μl Taq DNA polymerase 0.2μL, 10μM upstream and downstream primers 0.6μL, 1×LC Green 1μL, template DNA 0.8μL, 5M Betain 2.0μL, DMSO 1.0μL, deionized water 3.0μL;
[0062] Step 3, add the DNA of the sample to be tested: add 1 μL of negative quality control products to the first three wells of the 96-well PCR plate, and add 1 μL of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com