Method for efficiently predicting Gram-negative bacteria type III and type IV effector proteins
A Gram-negative bacteria and effector protein technology, applied in proteomics, neural learning methods, biological neural network models, etc., can solve the problems of high cost, time-consuming, low prediction accuracy of effector proteins, etc., and reduce parameters Space, improve generalization ability, improve prediction efficiency and accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0044] In order to make the purpose, technical solutions and advantages of the present invention clearer, the technical solutions in the embodiments of the present invention will be clearly and completely described below in conjunction with the drawings in the embodiments of the present invention.
[0045] A method for efficiently predicting Gram-negative bacteria type III and type IV effector proteins, specific steps:
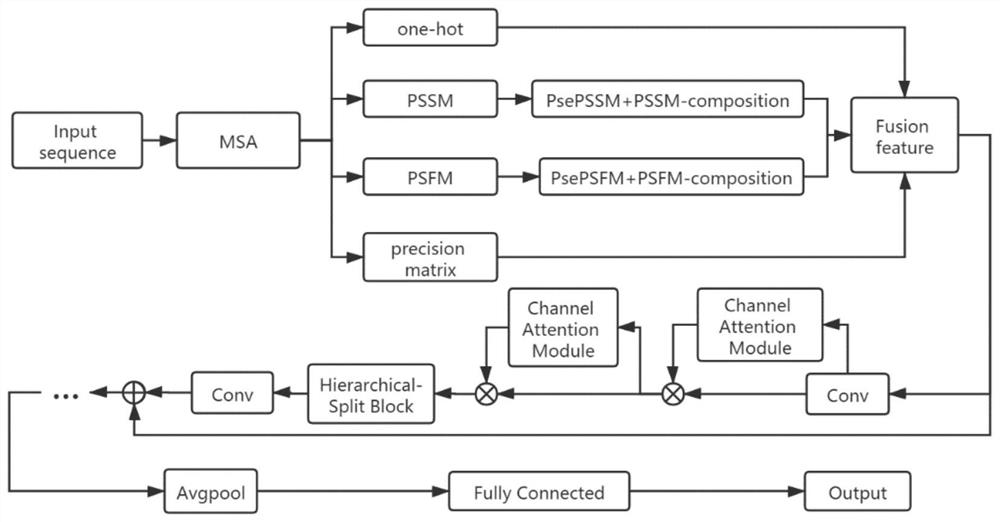
[0046] (1) Feature processing: multiple sequence alignment (MSA) was generated by HHsuite (version 3.0.1), and the database searched by HHsuite was UniRef30 (version 2020_03). Input the T3SEs and T4SEs sequences into the HHsuite program, and then use the HHsuite program to search for homologous sequences in the protein sequence library, and then construct the MSA, and then calculate the one-hot encoding and position-specific score matrix (PSSM) from the MSA , position-specific frequency matrix (PSFM) and precision matrix.
[0047] The model adopts the attenti...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com