Pretreatment method of RNA enzymatic hydrolysate
A technology of pretreatment and enzymatic hydrolysis, which is applied in the field of pretreatment of RNA enzymatic hydrolysis, can solve problems such as poor separation effect, achieve the effects of prolonging life, uniform distribution, and increasing exchange capacity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Take the reaction solution 001, see Table 1:
[0030] name A430 Concentration C Adsorption capacity Remark Reaction solution 001 1.083 32.75 1 Volume 3L
[0031] Table I
[0032] Reaction solution 001 is realized at (molecular weight 3000D, pump pressure 0.2Mpa), (molecular weight 6000D, pump pressure 0.2Mpa), (molecular weight 10000D, pump pressure 0.2Mpa), (molecular weight 20000D, pump pressure 0.2Mpa) , and get the following data, see Table 2:
[0033] molecular weight pump pressure A430 Concentration C time Adsorption capacity exchange adsorption increase Separation 3000D 0.2Mpa 0.390 25.29 50min 1.18 18% improve 6000D 0.2Mpa 0.455 28.35 42min 1.13 13% improve 10000D 0.2Mpa 0.622 29.58 30min 1.08 8% improve 20000D 0.2Mpa 0.921 30.12 24min 1.05 5% no change
[0034] Table II
[0035] In summary, through the ultrafiltration membranes with different mol...
Embodiment 2
[0038] The filtrate enters the ultrafiltration machine, and the membrane molecular weight of the ultrafiltration machine is set to 3000D, and the pump pressure is 0.2Mpa for ultrafiltration, and ultrafiltration is performed for 10, 20, 30, 40, and 50 minutes respectively, and samples are taken within the ultrafiltration time range Ultrafiltrate and effluent, detect the ultraviolet absorption wavelengths A250, A260, A280, A430 of the filtrate, ultrafiltrate and effluent, and calculate the ratio A250 / A260 (ratio 1) and A80 / A260 (ratio 2) and Concentration C;
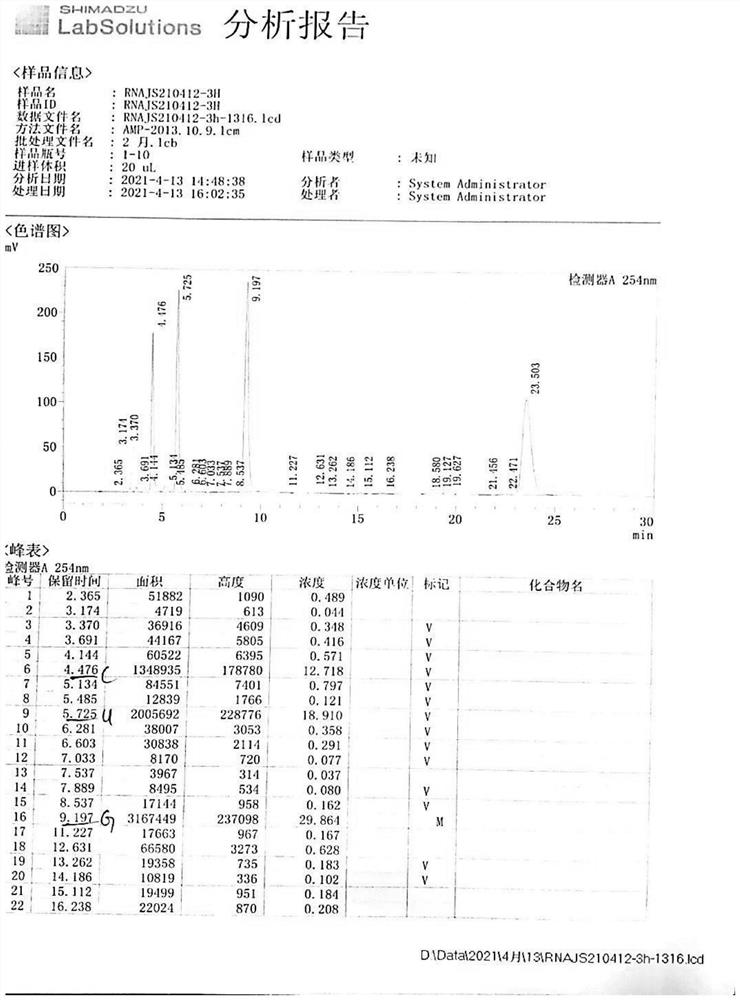
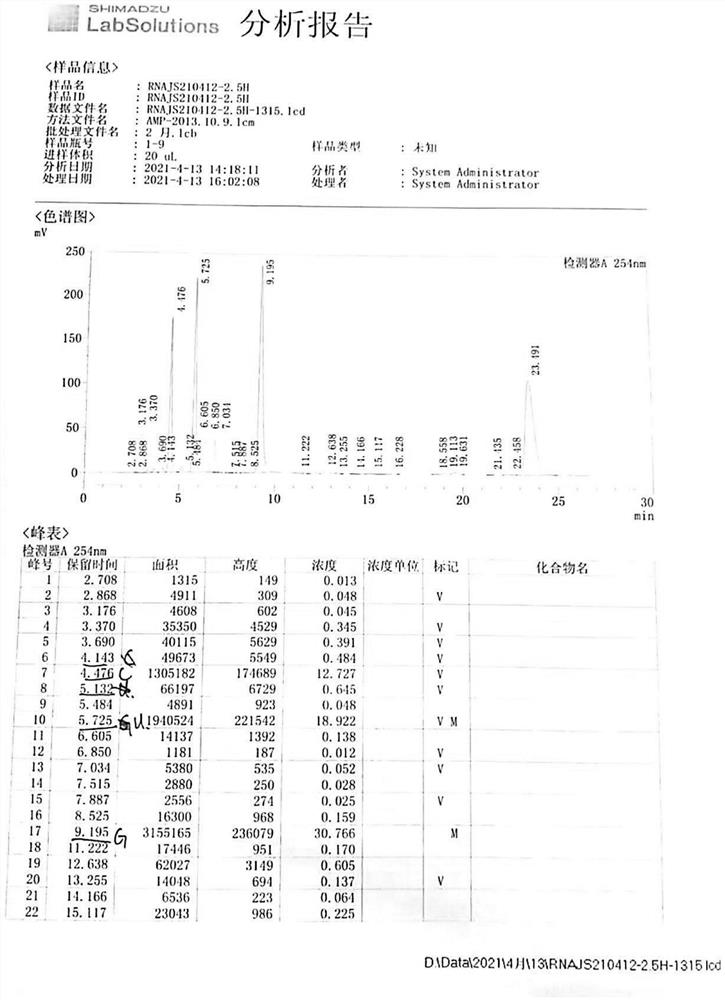
[0039] RNA reaction liquid ultrafiltration (taking 3000 molecular weight as an example), see Table 3:
[0040] name A250 A260 A280 Ratio 1 Ratio 2 Concentration C A430 Reaction solution 002 0.405 0.483 0.307 0.84 0.64 32.75 1.083 Ultrafiltration 10min 0.670 0.805 0.502 0.83 0.62 27.29 0.481 Ultrafiltration 20min 0.335 0.401 0.252 0.84 0.63 27.19 0.517 Ultrafiltra...
Embodiment 3
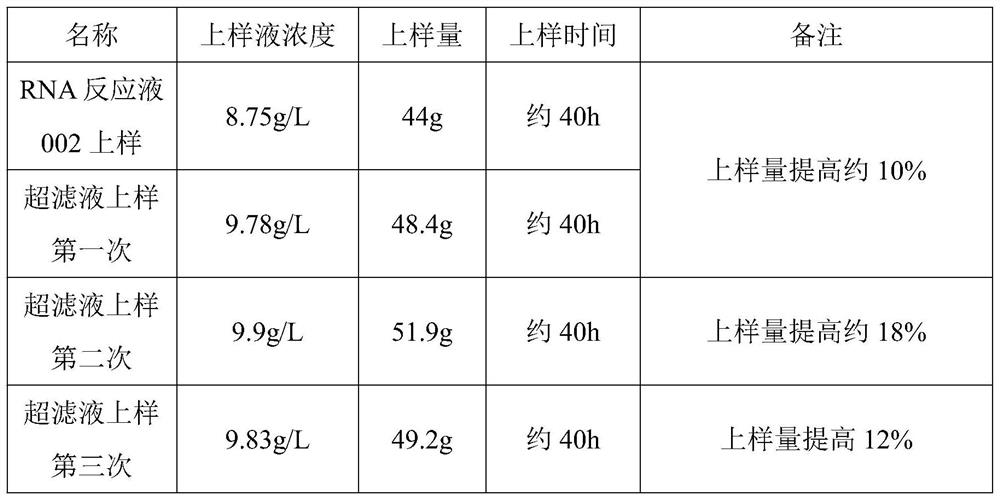
[0044] Load the ultrafiltrate and the RNA reaction solution separately, if the leakage concentration C≥0.4g / L, and the ratio of the concentration of the sample solution to the concentration of the leakage solution is 2≥1, it is judged as a sample leakage, record the concentration of the sample and the concentration of the sample. amount, and sample loading time, etc., and calculate the amount of sample loading increase, see Table 4:
[0045]
[0046] Table four
[0047] To sum up, the increase in ultrafiltration loading of the RNA reaction solution in the small test was greater than 10%, and the maximum increase was 18%.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com