Ketolytic acid hydroxymethyltransferase mutant, coding gene and application of mutant
An acid hydroxymethyl, transferase technology, applied in transferase, application, genetic engineering and other directions, can solve the problems of low yield, increased pan-operon transcription, high production cost, and achieve the effect of clear structure
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
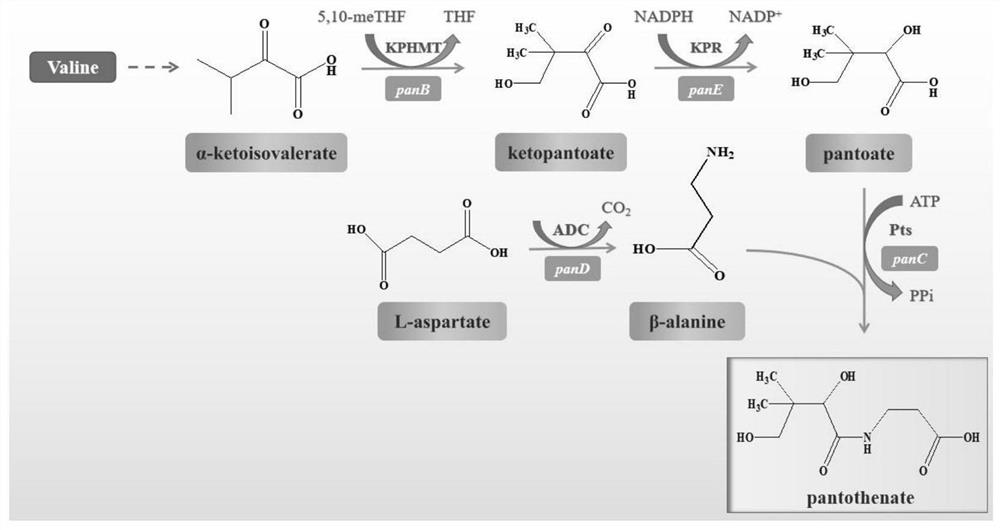
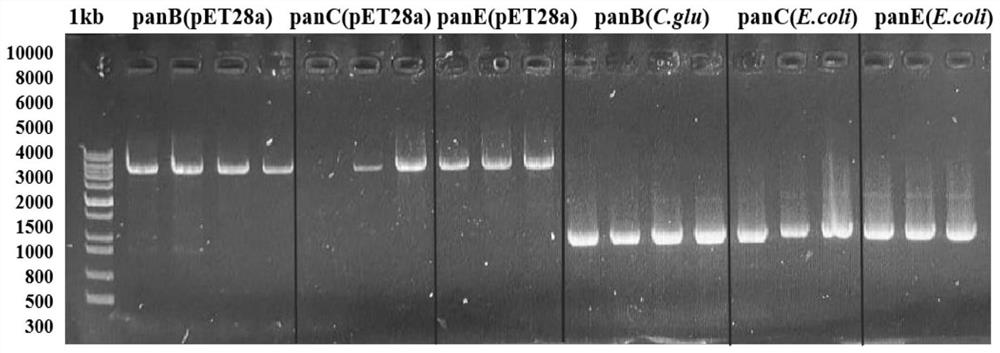
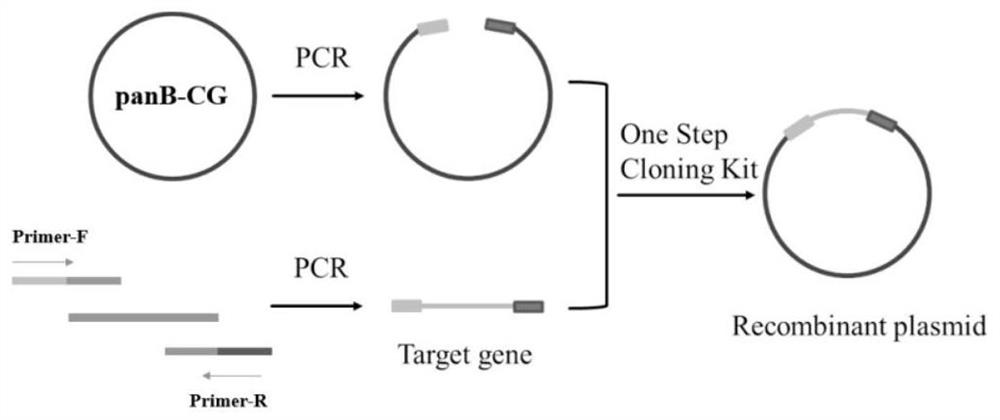
[0038] Example 1: Construction of wild-type ketopantoate hydroxymethyltransferase genetically engineered bacteria panB-CG
[0039] The gene sequence of Corynebacterium glutamicum ATCC 13032 ketopantoate hydroxymethyltransferase KPHMT (GenBank accession number: BX927148.1) in the gene bank was obtained by total gene synthesis to obtain the panB-CG plasmid. Design expression primer 1 (tttgtttaactttaagaaggagatataccATGCCCATGTCAGGCATTGATGCAAAG), primer 2 (tctcagtggtggtggtggtggtgctcgagAAAGGACTCCGCTTCGCCTGGGAAGGT), using Max high-fidelity DNA polymerase was used for PCR amplification to obtain the 813bp ketopantoate hydroxymethyltransferase gene sequence (the amino acid sequence is shown in SEQ ID NO.1, and the nucleotide sequence is shown in SEQ ID NO.2). Using the pET28a vector as a template, the linearized vector was obtained by PCR amplification technique. Primers were designed as follows, primer 3 (ACCTTCCCAGGCGAAGCGGAGTCCTTTctcgagcaccaccaccaccaccactgaga), primer 4 (CTTTGCATCA...
Embodiment 2
[0040] Example 2: Construction of wild-type ketopantoate reductase gene engineering bacteria panE-EC
[0041] The gene sequence of Escherichia coli str. K-12substr, W3110 ketopantoate reductase KPR (GenBank accession number: BAE76205.1) in the gene bank was obtained by total gene synthesis to obtain the panE-EC plasmid. Design expression primer 5 (tttgtttaactttaagaaggagatataccATGAAAATTACCGTATTGGGATGCGGTGCC), primer 6 (tctcagtggtggtggtggtggtgctcgagCTACCAGGGGCGAGGCAAACCAGTGCCGAT), using Max high-fidelity DNA polymerase performs PCR amplification to obtain a 912bp ketopantoate reductase gene sequence (the nucleotide sequence is shown in SEQ ID NO.3, and the amino acid sequence is shown in SEQ ID NO.4). Using the pET28a vector as a template, the linearized vector was obtained by PCR amplification technique. Primers were designed as follows, primer 7 (ATCGGCACTGGTTTGCCTCGCCCCTGGTAGctcgagcaccaccaccaccaccactgaga), primer 8 (GGCACCGCATCCCAATACGGTAATTTTCATggtatatctccttcttaaagttaaacaa...
Embodiment 3
[0042] Example 3: Construction of wild-type pantothenate synthetase genetically engineered bacteria panC-EC
[0043] The gene sequence of pantothenate synthase Ps derived from Escherichia coli str. K-12substr, W3110 in the gene bank (GenBank accession number: BAE76042.1) was obtained by total gene synthesis to obtain the panC-EC plasmid. Design expression primer 9 (tttgtttaactttaagaaggagatataccATGTTAATTATCGAAACCCTGCCGCTGC), primer 10 (tctcagtggtggtggtggtggtgctcgagTTACGCCAGCTCGACCATTTTGTTGTCGAT), using Max high-fidelity DNA polymerase performs PCR amplification to obtain a 855bp pantothenate synthetase gene sequence (the nucleotide sequence is shown in SEQ ID NO.5, and the amino acid sequence is shown in SEQ ID NO.6). Using the pET28a vector as a template, the linearized vector was obtained by PCR amplification technique. Primers were designed as follows, primer 11 (ATCGACAACAAAATGGTCGAGCTGGCGTAActcgagcaccaccaccaccaccactgaga), primer 12 (GCAGCGGCAGGGTTTCGATAATTAACATggtatatctc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com