Rice tryptophan decarboxylase and production method thereof
A technology of tryptophan decarboxylase and rice, which is applied in the field of bioengineering, can solve the problems of high price and achieve the effect of increasing the expression level and increasing the expression level
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Embodiment 1: Cloning and codon optimization of rice tryptophan decarboxylase (TDC) gene
[0031] The leaves of nutrient-deficient rice were selected, and the total RNA of the material was extracted by the CTAB method, according to the GenBank EST database ( https: / / www.ncbi.nlm.nih.gov / genbank / GenBank The complete cDNA ORF sequence information obtained in ) was used to design primers (TDCsense and TDCantisense), and the cDNA synthesized by reverse transcription was used as a template to amplify the full-length open reading frame by PCR.
[0032] TDCsense: 5'-GTAAGGTATAGTATCATC-3'; (SEQ ID NO.4)
[0033] TDCantisense: 5'-GTGAGTTAGCTAGGTTGGTGTG-3'. (SEQ ID NO.5)
[0034] The full-length cDNA of the rice tryptophan decarboxylase (TDC) gene includes a complete open reading frame of 1533bp, and the nucleotide sequence is shown in SEQ ID NO.3; it encodes a polypeptide of 507 amino acids, and its theoretical molecular weight is 56.21ku.
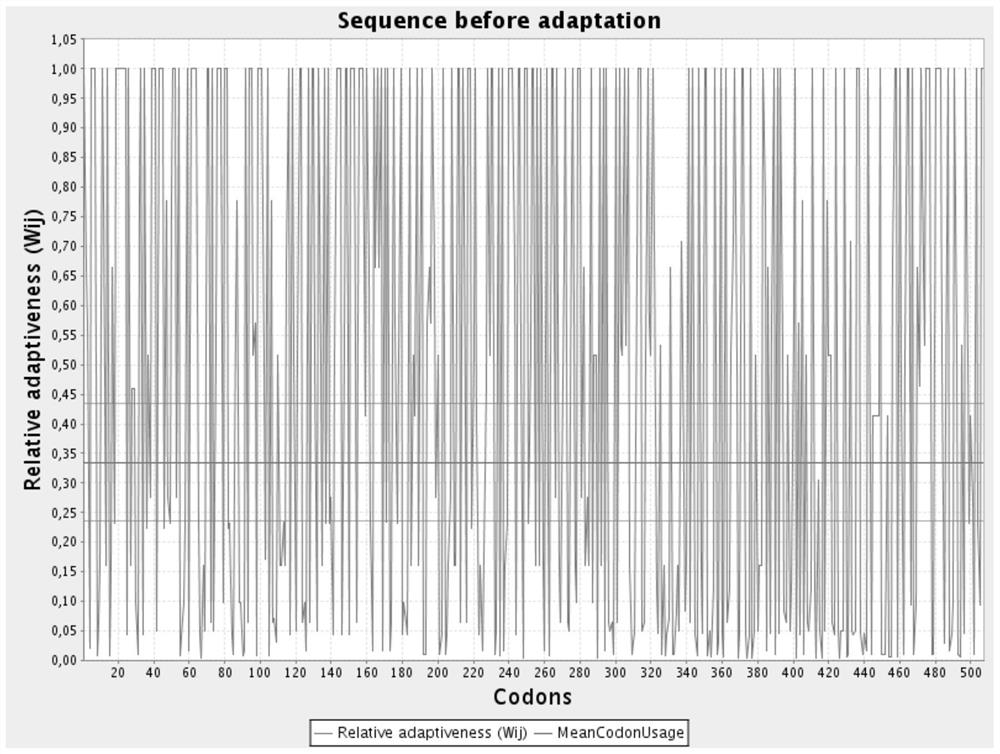
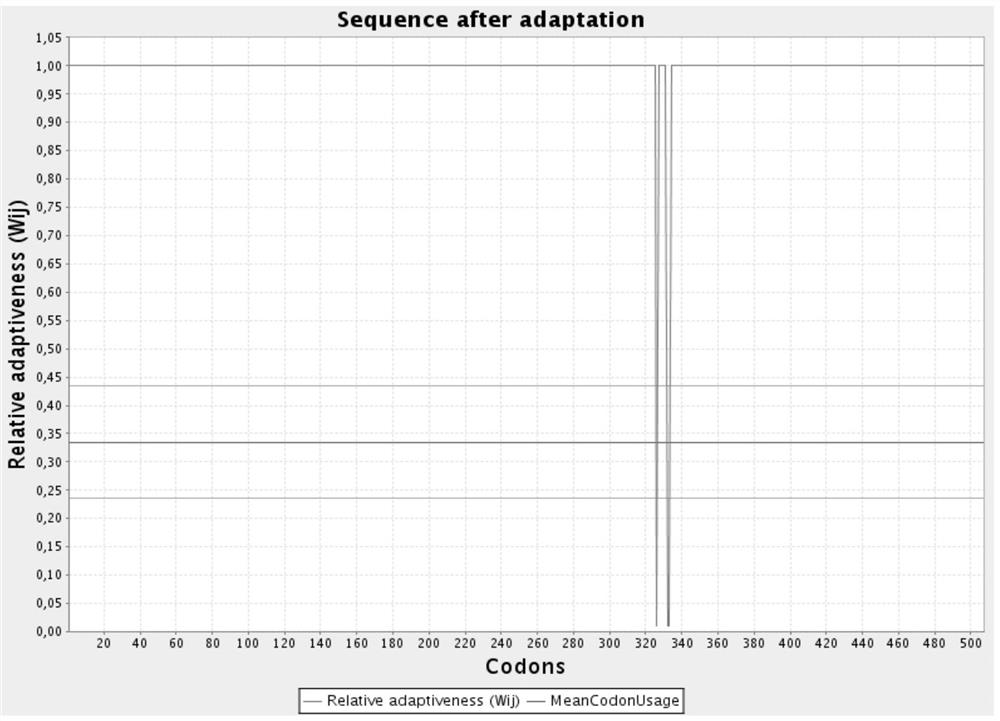
[0035] In order to make the gene ...
Embodiment 2
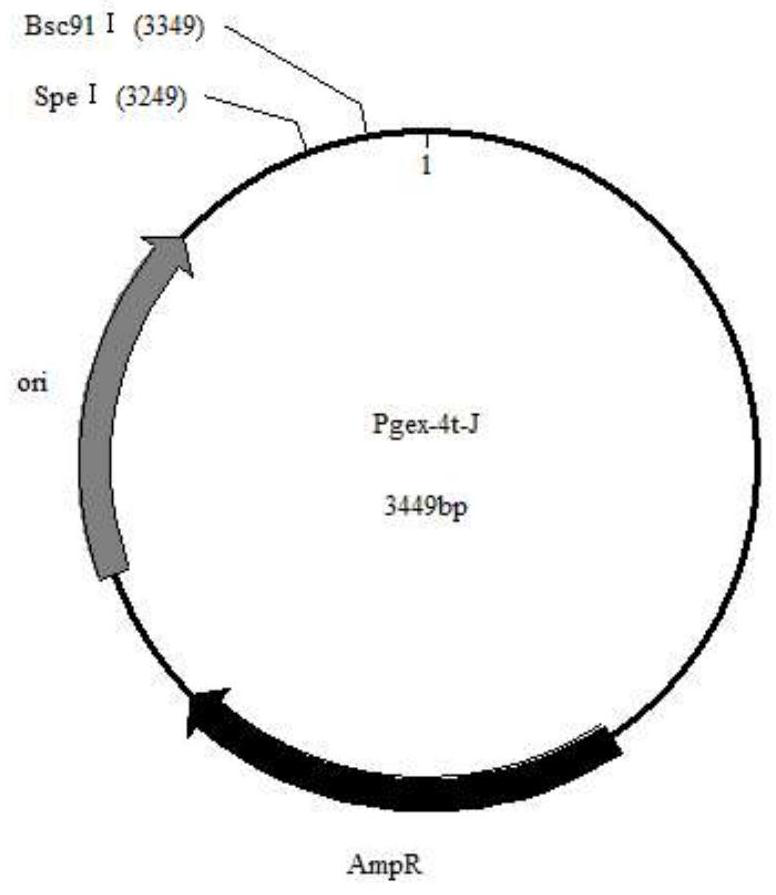
[0037] Embodiment 2: Construction of expression vector pGEX-4t-J:
[0038] Using the plasmid pGEX-4T-1 as the starting plasmid, first use pflmI to carry out a single enzyme cut at the 3250 position, then cut 2 pieces at the 3' end, cut 6 pieces at the 5' end, add A to the 3' end, and add CTAGT to the 5' end ;Reprotect; then use btgI to perform single enzyme digestion at the 4869 position, cut 4 pieces at the 5' end and then add AAGA to remove all protection; plus 5'-CTAGT...G-3' full-length 100bp Artificial chain, the omitted sequence in the middle of the artificial chain, as long as it does not contain the two enzyme cutting sites of SpeI and Bsc91I, the omitted sequence in the middle can be selected arbitrarily.
[0039] Plasmid pGEX-4t-J was double-digested with SpeI and Bsc91I and identified. After double-digestion, pEGX-4t-J appeared 3349 and 100, which proved that the expression vector pGEX-4t-J was successfully constructed.
Embodiment 3
[0040]Embodiment 3: Fermentative production of rice tryptophan decarboxylase
[0041] (1) Use SpeI and Bsc91I to double-digest the plasmid pGEX-4t-J, and then integrate the optimized TDC gene shown in SEQ ID NO.1 into the double-digested expression vector pGEX-4t by DNA ligase -J to obtain the recombinant expression vector pEGX-TDC; and then introduce the obtained recombinant expression vector into Escherichia coli B21 (DE3) to obtain the recombinant bacteria.
[0042] (2) Inoculate the recombinant bacteria into the culture medium for fermentation and cultivation. The conditions for fermentation and cultivation are: pH control at 7.0, tank pressure at 0.05MPa, temperature at 33°C, ventilation ratio 1:1, and fermentation until the fermentation medium is diluted 100 times. After OD 600 Value is 0.18-0.20, obtains fermentation medium;
[0043] Cool the fermentation culture liquid to 22°C, add the inducer IPTG to the fermentation culture liquid to make the final concentration of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com