Novel single-celled spatial transcriptome technology for tissue
A space transcription, single-cell technology, applied in the field of single-cell space transcriptome technology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
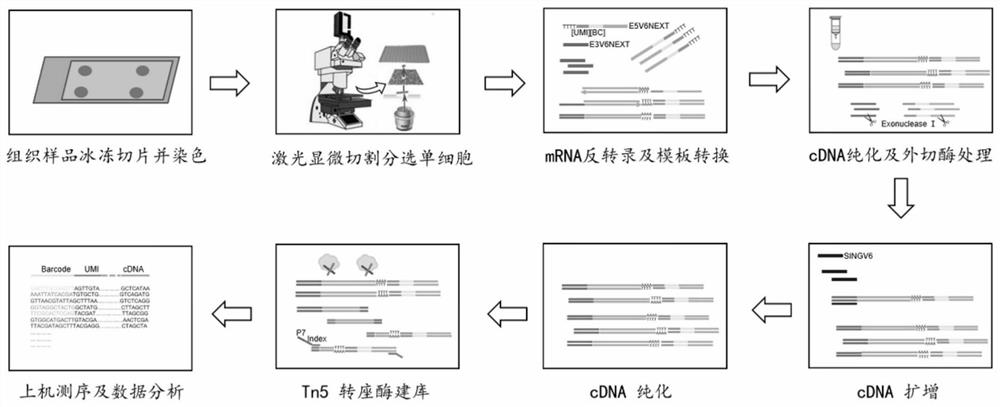
[0131] LCM-SCRB-seq method of the present invention
[0132] 1. Preparation of Cell Collection Solution
[0133] The buffer solution (containing Phusion HF buffer diluted 500 times, 2U / μL RNase inhibitor, 0.2% Triton X-100) was prepared according to the following composition.
[0134] Table 1
[0135] Reagent volume Phusion HF buffer (New England Biolabs, USA) 2μL RNase Inhibitor 35.8μL 2% Triton X-100 71.6μL Nuclease-free Water 606.6μL total capacity 716μL
[0136] Add 10 μL of cell collection solution (8 μL of buffer and 2 μL of cell-tagged primer (E3V6NEXT, US14898030A1) at a concentration of 2 μM) to a single nuclease-free PCR tube, or inside the lid of an LCM 96-well collection plate Add 7 μL of cell collection solution (5 μL of buffer and 2 μL of cell-labeled primers at a concentration of 1 μM) to each well, cover the 96-well plate, and place the plate with the cover down and put it in a 4°C refrigerator for later use....
Embodiment 2
[0188] Characteristic analysis of the LCM-SCRB-seq method of the present invention
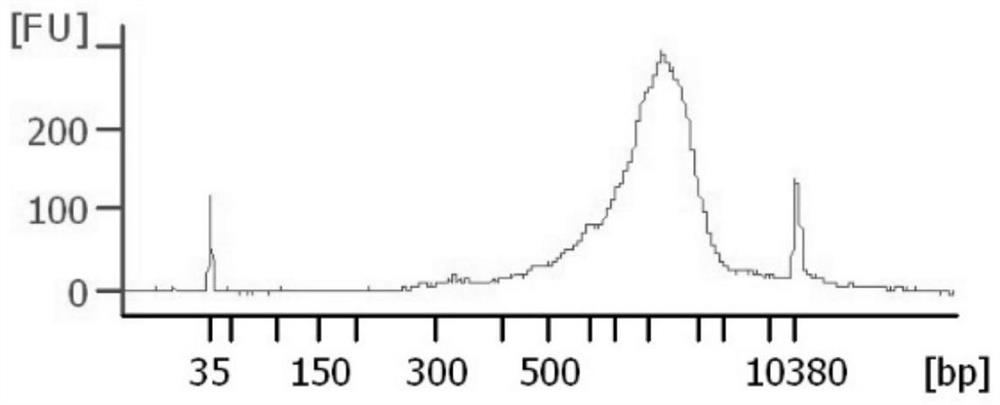
[0189] 1. Quality detection of cDNA in single cells captured in situ from tissues
[0190] As described in Example 1, the LCM-SCRB-seq method of the present invention was used to sort single cells by laser microdissection, mRNA inversion, cDNA amplification and purification, and the obtained cDNA was analyzed by Agilent 2100 for quality inspection.
[0191] figure 2 The quality inspection results show that the full-length cDNA obtained by the LCM-SCRB-seq method of the present invention accounts for more than 50% of the length of more than 1000bp, and even exceeds about 50% of the single-cell transcriptome gold standard SMART-seq2. Database building requirements.
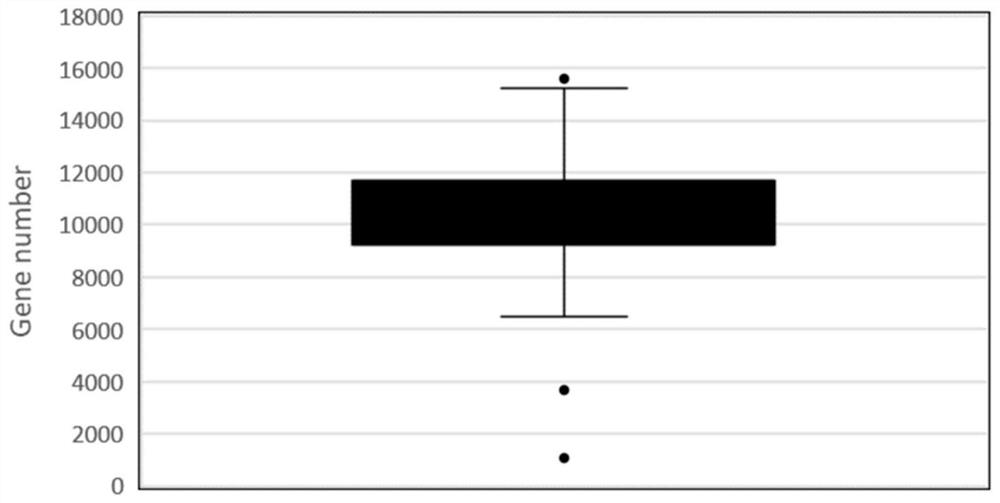
[0192] 2. Sensitivity detection
[0193] Using the LCM-SCRB-seq method of the present invention (as described in Example 1), the oocytes of 30-day normal mice were constructed and sequenced.
[0194] The results showed that at th...
Embodiment 3
[0199] Single-cell transcriptome sequencing of 30-day mouse oocytes using the LCM-SCRB-seq method of the present invention
[0200] 1. Preparation of Cell Collection Solution
[0201] Add 7 μL of cell collection solution (5 μL of buffer and 2 μL of cell-labeled primers at a concentration of 1 μM) to each well in the cover of the LCM 96-well collection plate. The buffer solution is prepared according to Table 1 in Example 1, and the 96 wells are covered Plates, with the plates covered, placed in a 4°C refrigerator for later use.
[0202] 2. Freezing section and staining
[0203] Take out the 30-day mouse ovarian tissue from -80°C, freeze the section at 20 μm, paste the tissue section on the filmed PEN slide that has been pre-cooled in ice, fix it in 95% ethanol for 2 minutes, and stain with methyl violet staining solution for 1 minute and 50 seconds , after drying with ethanol gradient dehydration, air-dry the slides naturally for a few seconds, and balance the temperature sl...
PUM
| Property | Measurement | Unit |
|---|---|---|
| thickness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com