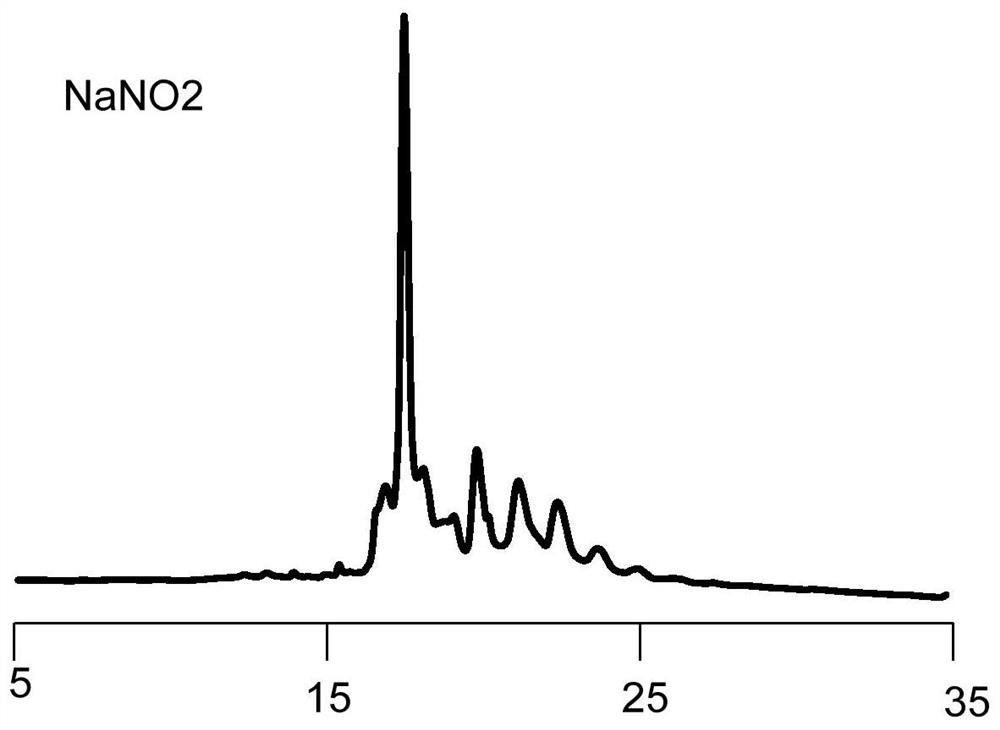
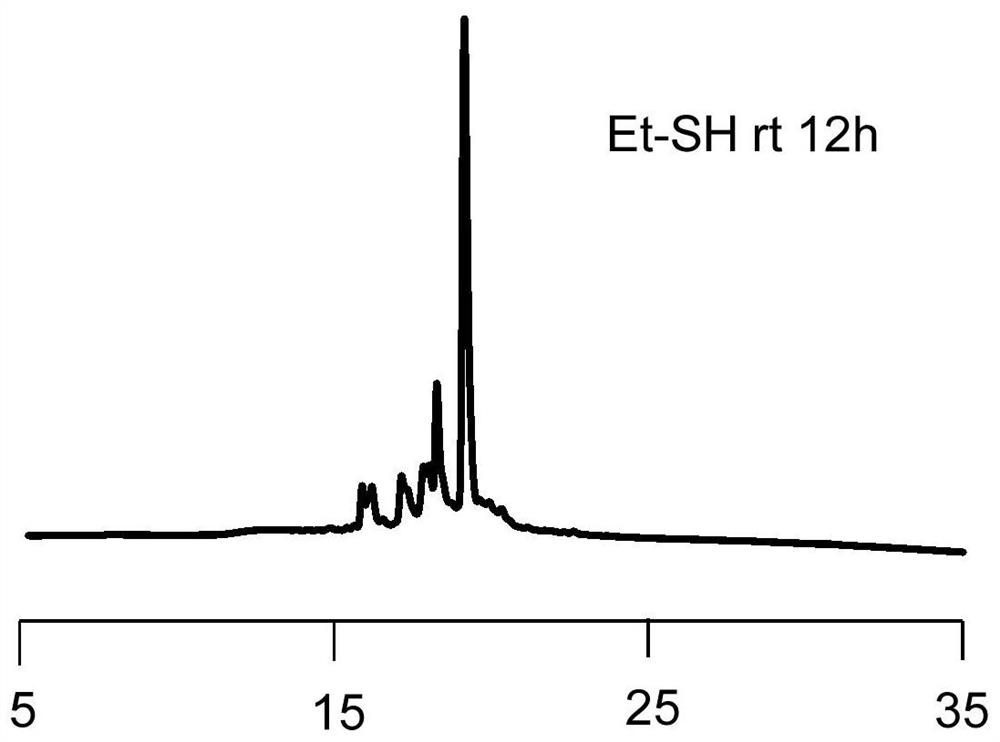
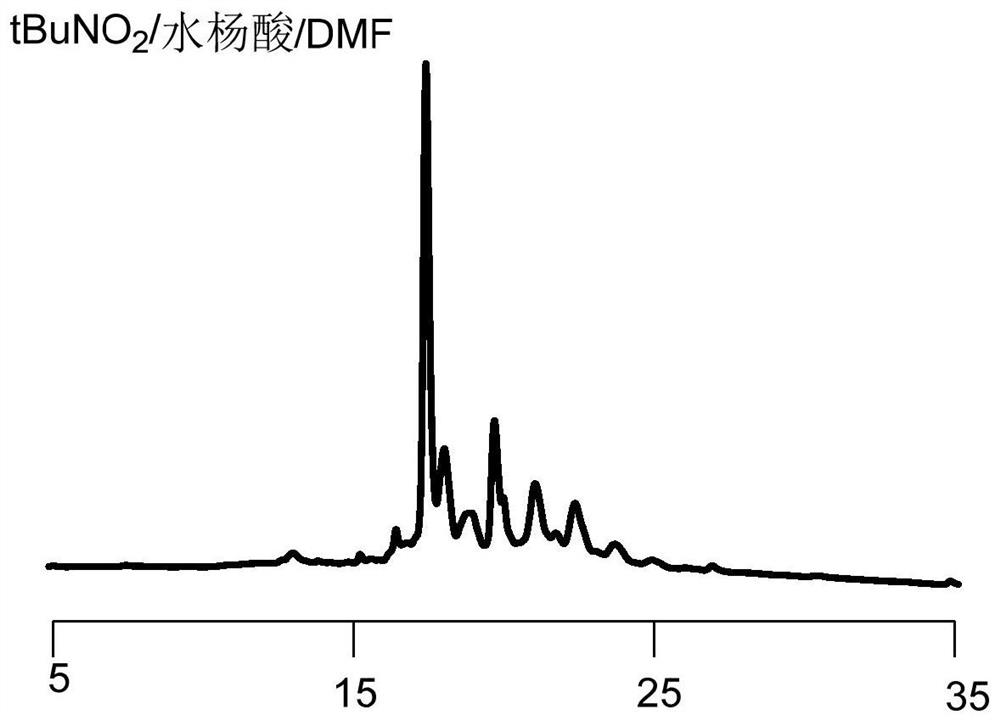
Novel method for rapidly removing removable framework modification and application
A new method and removal technology, applied in the field of preparation of polypeptides or proteins, can solve the problems of difficult removal and slow removal, and achieve the effects of reducing aggregation, inhibiting the formation of secondary structures, and high yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0087] The fourth aspect of the present invention provides a method for preparing a polypeptide, comprising the following steps:
[0088] S1, reacting 2-hydroxy-4-methoxy-5 nitrobenzaldehyde with the amino group at the N-terminal of the polypeptide on the solid-phase resin to generate a carbon-nitrogen double bond structure to obtain the first polypeptide compound. Since it is a prior art, no longer elaborate;
[0089] S2. In an organic solution, reducing the first polypeptide compound with a reducing agent to reduce the carbon-nitrogen double bond structure to a carbon-nitrogen single bond structure to obtain a second polypeptide compound, the purpose of which is to reduce the introduction of RBM carbon-nitrogen double bond, in this step, the organic solution can be a conventional choice, such as DMF solution, the reducing agent here is not particularly limited, can be a conventional choice in the art, specific examples include but are not limited to NaBH 4 , sodium triaceto...
Embodiment 1
[0096] This example discloses a method for preparing a model peptide, comprising the following steps:
[0097] S1, contacting 2-hydroxy-4-methoxy-5 nitrobenzaldehyde with the polypeptide on the solid phase resin shown in formula (a1), to obtain the polypeptide on the solid phase resin shown in formula (a2),
[0098]
[0099] Wherein, the solid-phase resin shown in formula (a1) is obtained by Fmoc solid-phase synthesis method, and the specific steps are: adding Rink AM resin to a polypeptide synthesis tube, and using N,N dimethylformamide (DMF) / dichloromethane ( DCM) (volume ratio 1:1) was swollen for 30 minutes, then 20% piperidine in DMF solution was added to treat the resin for 5 minutes and 10 minutes, and the resin was washed three times with DMF, DCM and DMF successively. Then 3.8eq of 2-(7-azobenzotriazole)-tetramethyluronium hexafluorophosphate (HATU), 8eq of N,N diisopropylethylamine (DIEA) and 4eq of Fmoc-protected The first amino acid at the C-terminal of the tar...
Embodiment 2
[0125] This example adopts the same implementation method as Example 1, except that the reaction temperature in step S52 is 0°C.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com