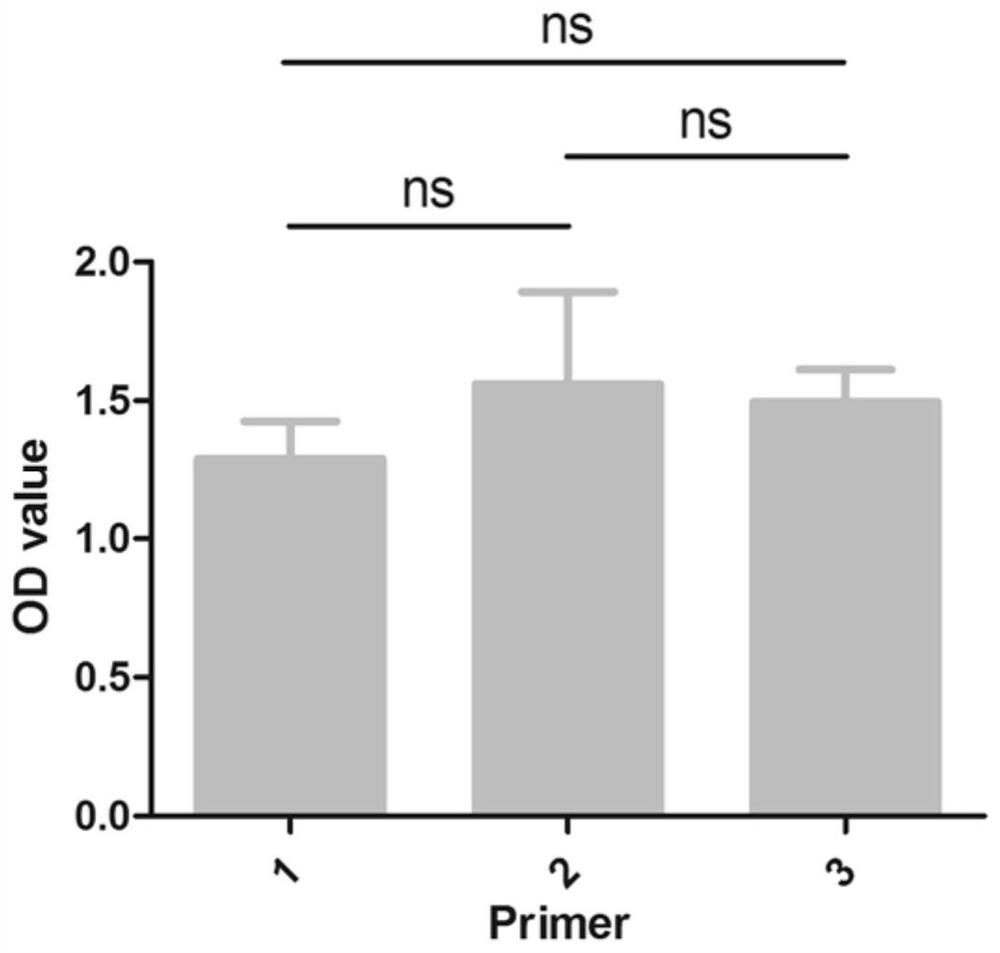
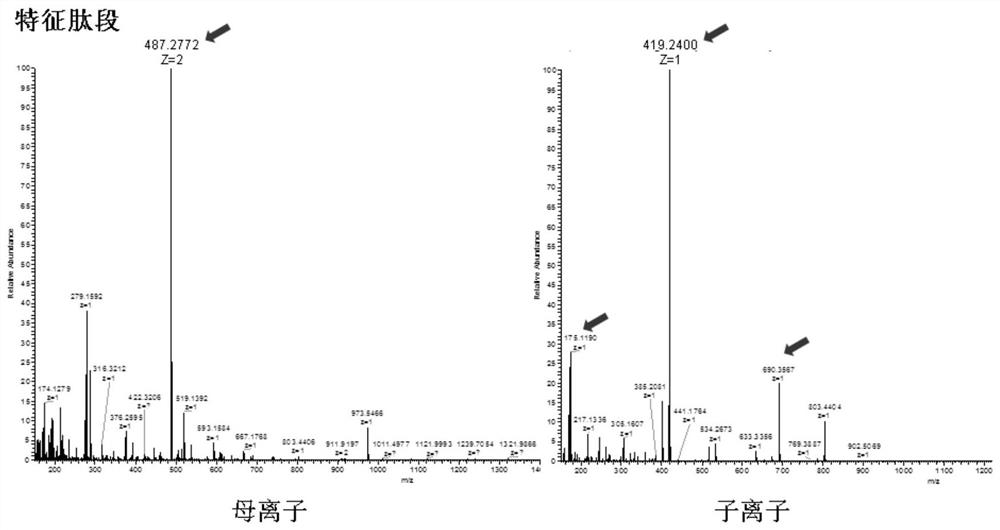
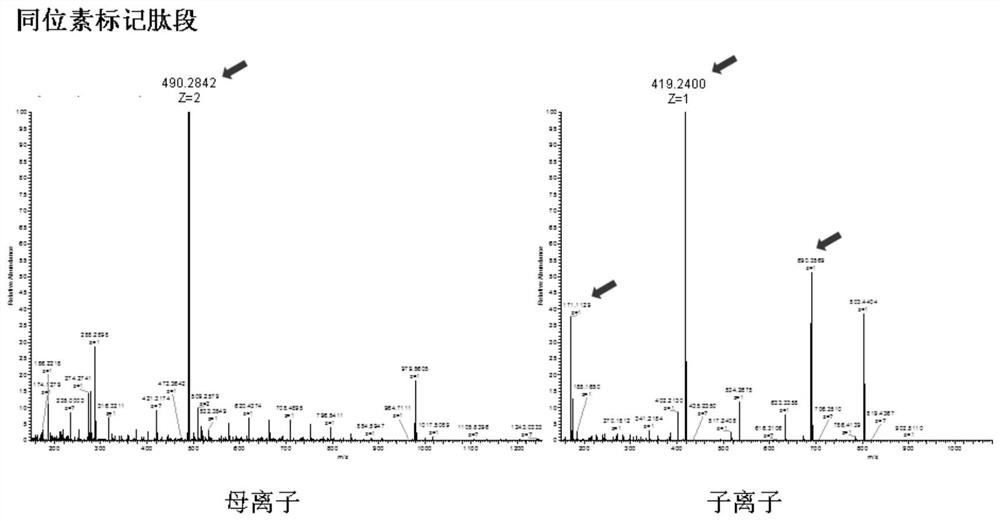
Mass spectrum method for quantifying nucleic acid based on DNA-polypeptide probe technology and application thereof
A technology of DNA probes and probes, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve problems affecting PCR amplification efficiency and response, deviation, and affecting signal collection, achieving high accuracy, High precision and high detection specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Embodiment 1 Construction of mass spectrometry method based on DNA-polypeptide probe technology quantitative nucleic acid
[0047] This embodiment takes HULC nucleic acid as an example, and provides a mass spectrometry method based on DNA-polypeptide probe technology to quantify HULC. Based on the nucleotide sequence of the nucleic acid to be tested, DNA probes are designed; characteristic polypeptides are screened out, and combined with DNA probe to construct a DNA-polypeptide probe; hybridize the DNA-polypeptide probe with the nucleic acid to be tested; perform proteolysis on the hybridized DNA-polypeptide probe, so that the characteristic polypeptide in the DNA-polypeptide probe is hydrolyzed into a reporter peptide ; Construct the LC-MS / MS detection method of the reporter peptide; use the LC-MS / MS detection method to detect the concentration of the reporter peptide; convert the concentration of the reporter peptide into the copy number of the nucleic acid to be teste...
Embodiment 2
[0220] Example 2 Application of the construction-based nucleic acid mass spectrometry quantitative method in the detection of HULC copy number in hepatocytes
[0221] In this embodiment, five different hepatocytes were used as samples to be tested, and the copy number of HULC was detected. The five kinds of liver cells used were normal liver cell L02, normal liver cell 7702, liver cancer cell SK-Hep1, liver cancer cell HepG 2 , Liver cancer cell 7721.
[0222] Refer to the construction method of the DNA-polypeptide probe in Example 1, the hybridization of the DNA-polypeptide probe and the nucleic acid to be tested, the enzymatic hydrolysis to obtain the characteristic polypeptide reporter peptide, and the LC-MS / MS of the characteristic polypeptide reporter peptide constructed based on Example 1 The detection method is to detect the copy number of HULC in the above five kinds of hepatocytes.
[0223] Wherein, the extraction process of total RNA in the above five kinds of live...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com