CRISPR/Cas12a one-step nucleic acid detection method
A detection method and nucleic acid technology, applied in the field of genetic detection, can solve problems such as prolonged detection time, false positives, and unfavorable practical application.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0022] In order to make the technical means and effects realized by the present invention easy to understand, the present invention will be described in detail below in conjunction with the embodiments and accompanying drawings.
[0023]
[0024] Reagents, instruments and bacterial strains used in this embodiment are as follows:
[0025] Main reagent: LbCas12a Guangzhou Bolaisi Biotechnology Co., Ltd.; XP Beckman Coulter; RPA Amplification Kit Weifang Anpu Future Biotechnology Co., Ltd.; Primer Sequence Sangon Bioengineering Co., Ltd.; HiScribe T7 Quick HighYield RNA Synthesis Kit New England Biolabs.
[0026] Main instruments: Microplate reader SpectraMax M2 was purchased from Molecular Devices; Nanodrop 2000C was purchased from Thermo Scientific; HiScribe T7 Quick HighYield RNA Synthesis KitNewEnglandBiolabs.
[0027] The strains used are shown in Table 1:
[0028] Table 1 Strain names and corresponding numbers
[0029]
[0030]
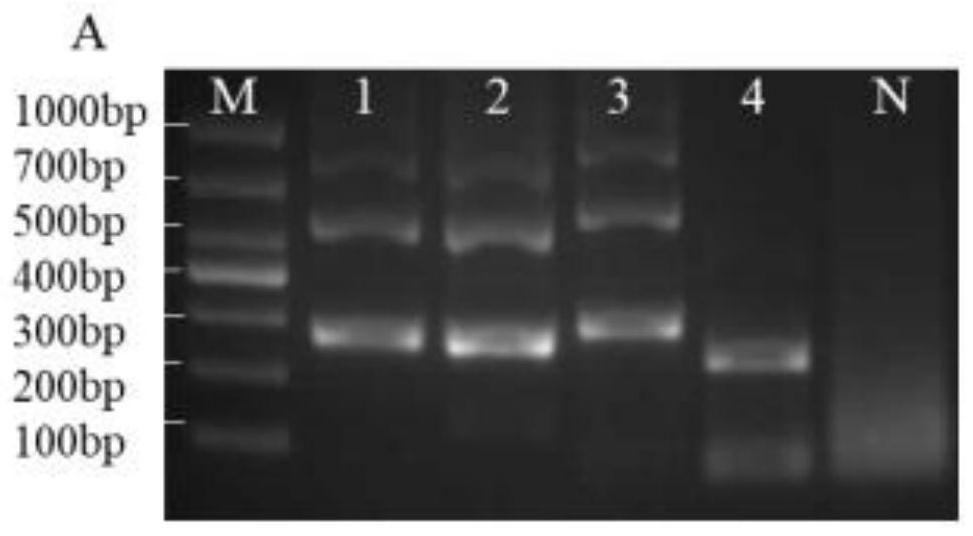
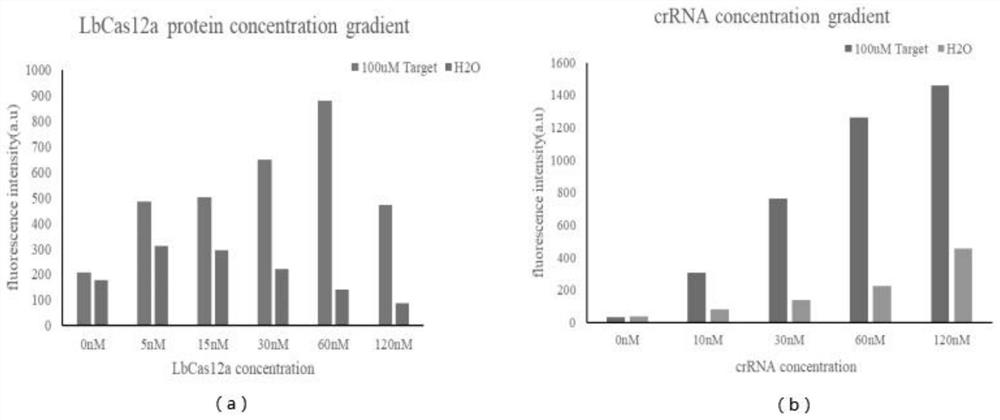
[0031] figure 1It is a schemat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com