A screening method for nucleic acid aptamer specifically binding to sarafloxacin hydrochloride
A technology of salafloxacin hydrochloride and nucleic acid aptamer, which is applied in the field of chemical analysis, can solve the problems of food safety consequences and antibiotic residues, and achieves the effects of short screening period, high affinity and convenient synthesis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0077] Embodiment 1: Synthesis of the ssDNA library and primers shown below:
[0078] Raw random ssDNA library:
[0079] 5'-AAGGAGCAGCGTGGAGGATA-N40-TTAGGGTGTGTCGTCGTGGT-3';
[0080] Upstream primer: 5'-AAGGAGCAGCGTGGAGGATA-3'
[0081] Downstream primer 1: 5'-ACCACGACGACACACCCTAA-3'
[0082] Downstream primer 2: 5'-bio-ACCACGACGACACACCCTAA-3'
Embodiment 2
[0083] Embodiment 2: SELEX technology screening specific nucleic acid aptamer
[0084] 2.1 The solution required for the experimental process
[0085] (1) 100mL selection buffer configuration method: 0.037g KCl, 0.011g CaCl 2 , 0.5844g NaCl, 0.019g MgCl 2 , 0.24g Tris, 20μL Tween 20, adjust the pH to 7.6, make up to 100mL with deionized water;
[0086] (2) 100mL washing buffer configuration method: 21g urea, 0.48g Tris, 0.37g EDTA. Na 2 , 20μL Tween 20, adjust the pH to 8.0, make up to 100mL with deionized water;
[0087] (3) 100mL Binding and Washing (BW) Buffer configuration method: 0.12g Tris, 0.037g EDTA. Na 2 , 11.68g NaCl, adjust the pH to 7.4, make up to 100mL with deionized water;
[0088] (4) 100mL amino magnetic bead coupling buffer configuration method: 1.95g 2-morpholineethanesulfonic acid, adjust the pH to 4.0, make up to 100mL with deionized water;
[0089] (5) 100mL amino magnetic bead blocking buffer configuration method: 2.4g Tris, adjust the pH to 8.0...
Embodiment 3
[0124] Example 3. High-throughput sequencing and sequence analysis
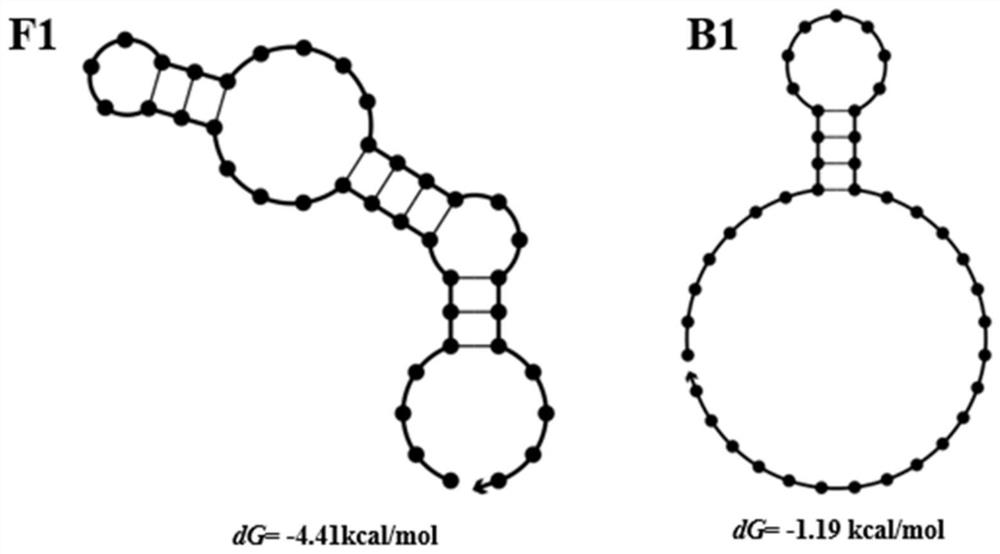
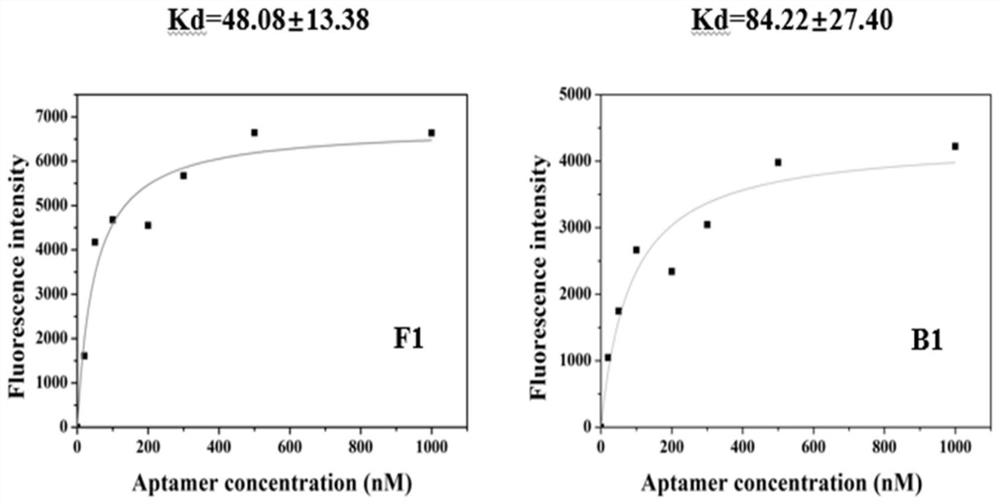
[0125] After the final round of screening products were amplified by PCR, they were subjected to agarose gel electrophoresis, and the 80bp bands were recovered by gel cutting, and then sent to the company for high-throughput sequencing. The sequence with the most occurrences in the high-throughput sequencing results is sent to the company for synthesis, and then the Kd value is tested, and the secondary structure of the sequence is predicted by software. The sequencing results exclude the formation of primer dimers and sequences containing the complementary strands of the upstream primer and the downstream primer 1. The two sequences with the smallest Kd and the highest affinity with the target molecule are as follows:
[0126] F1: AAGGAGCAGCGTGGAGGATACTCCGTGCGATCGCCGGGGACCGAAGAATCGTTCACATCGTTAGGGTGTGTCGTCGTGGT
[0127] B1: AAGGAGCAGCGTGGAGGATACCATCCACCTAGCATCCATAGGCGAACACTTTCTTGGGGCTTAGGGTGTGTCGTCGTGGT
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com