Primer pair, kit and method for detecting and identifying Pectobacterium bacteria
A pectin bacillus and primer pair technology, applied in the biological field, can solve the problems of low specificity, prone to false positive or false negative detection, low versatility, etc., and achieve the effect of accurate and sensitive inspection and quarantine
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032]Designed for general specific primers for the Pectobacterium strand 23S rRNA gene:
[0033]Download Pectobacteriaum in Pectobacteriacea, Pectobacteriacea, Pectobacteriacea, Pectobacteriacea, Pectobacteriacea, DICKEYA, Brenneria, and Lonsdalea, Erwinia and Pantoea in Owenia, Kosakonia in E. coli, and Klebsiella are complete or Close to complete (>The 2600 bp) 23S rRNA gene sequence, the nucleic acid sequence is collected with the MUSCLE program in Mega5 software, and the 23S rRNA gene of Typical P.carotovorum Typical strain NCPB 312 (= CFCC 10814) is a reference sequence, and the BioEdit software is displayed. Sequence Difference nucleotide site (figure 1 ), With 20 to 30 nucleotides as modules, scan sequences, found that there are multiple nucleotide differences in the Pectobacterium straighteners in the bacterial 23S rRNA gene sequence, which is a variety of nucleotide differences, which are sequences for these Pectobacterium specific conserved area sequences. , Design primers w...
Embodiment 2
[0041]Comparison primers were specific to PECTOBACTERIUM bacteria for 23Specf / 23SPECR, PEC-1F / PEC3R and PCC3F / PCC3R.
[0042]Each primer nucleotide sequence is as follows:
[0043]Positive primer 23specf: 5'-cggtaagctgattructgaagcgttatac-3 ',
[0044]Reverse primers 23SPECR: 5'-TGTGTACGGACTATCCCCCTTACTACT-3 ';
[0045]Positive primers PEC1-F: 5'-gtgcaagcgttaatcgaatg-3 ',
[0046]Reverse primers PEC1-R: 5'-ctctacaagactctcctgtcagtttt-3 ';
[0047]Positive primer PCC3F: 5'-gggattcgaaaaattactggctg-3 ',
[0048]Reverse primers PCC3R: 5'-gcttttctttcatcaacca-3 '.
[0049]Positive primer 23Specf and reverse primer 23SpeCr correspond to the nucleotide sequence of the 23S rRNA gene of Pectobacterium and other near-belly bacteria. The 23S rRNA gene sequence is displayed with the identical nucleotide of the primer corresponding to the nucleotide of the primer, and the nucleotide different from the corresponding nucleotide is shown in the form of a nucleotide (figure 1 ).figure 1 Showing reverse primers 23SPECR h...
Embodiment 3
[0055]The comparative primer was used to detect the sensitivity of the Pectobacterium bacteria in 23SPECF / 23SPECR and Y1 / Y2.
[0056]Each primer nucleotide sequence is as follows:
[0057]Positive primer 23specf: 5'-cggtaagctgattructgaagcgttatac-3 ',
[0058]Reverse primers 23SPECR: 5'-TGTGTACGGACTATCCCCCTTACTACT-3 ';
[0059]The forward primer Y1: 5'-ttaccgacgccgagctgtgggct-3 ',
[0060]Reverse primers Y2: 5'-caggaagatgtcgttatcgcgagt-3 '.
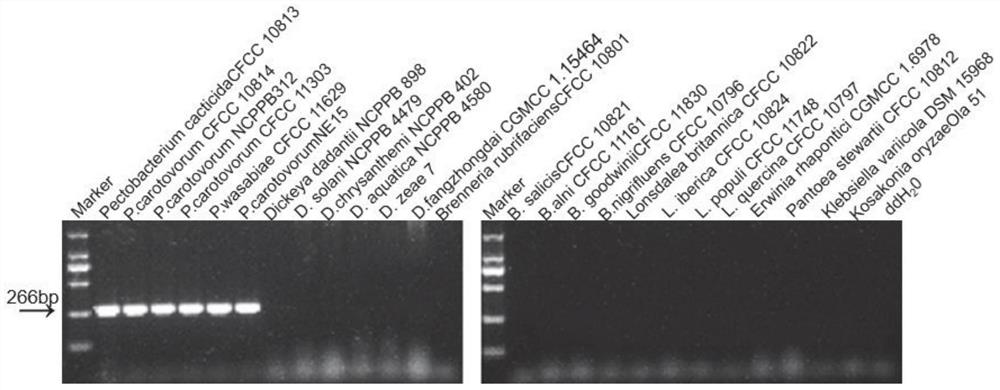
[0061]The Pectobacterium Carotovorum CFCC 10814 genomic DNA with a concentration of 40 ng / μl was diluted in 10 times series, and each of the 1 μl of DNA solution was a template, 23 JPECF and 23SPECR were used in 25 μl of reaction system, and Shanghai Hoheng Biotechnology is limited. The 2 × HIEFF PCP MASTER MIX produced by the company was carried out by the amplification procedure, and the PCR product (266 bp) was detected by agarose gel electrophoresis, showing the detection of the minimum concentration of 400 Fg / μL (Figure 6 ).
[0062]The Pectobacterium C...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com