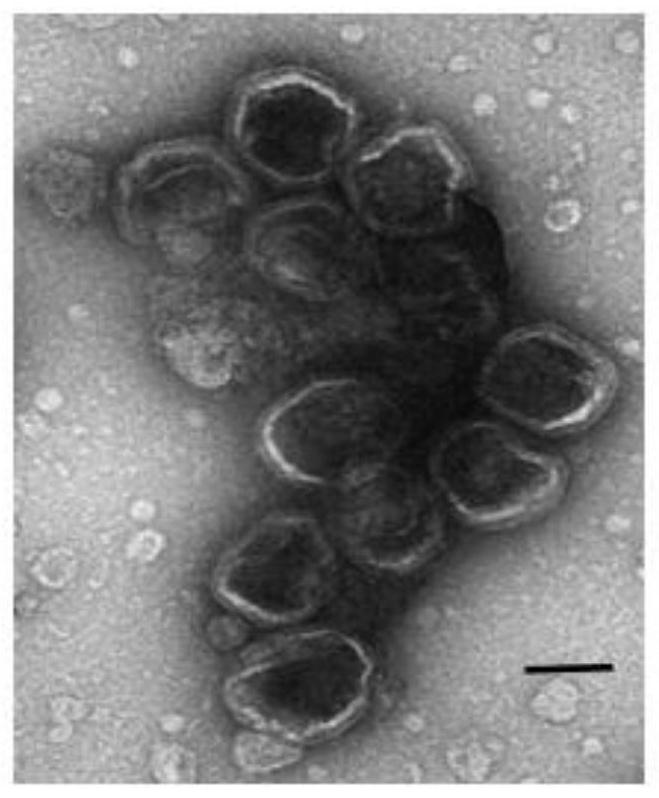
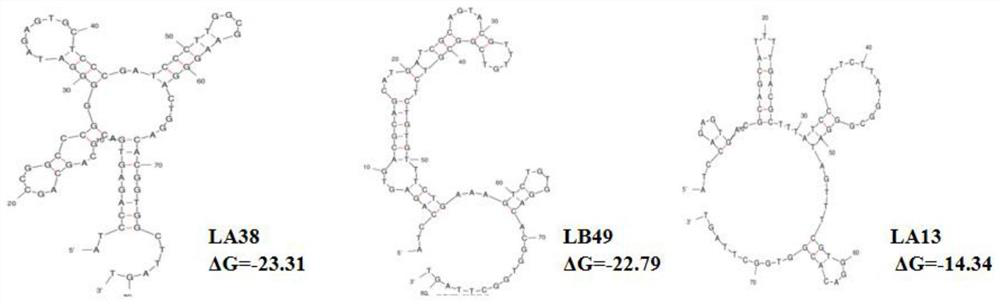
SsDNA aptamer for identifying micropterus salmoides iridovirus and application thereof
A nucleic acid aptamer and perch rainbow technology, which is applied in DNA preparation, recombinant DNA technology, DNA/RNA fragments, etc., can solve the problems of lack of rapid detection methods for LMBV and few research reports, and achieve high library enrichment efficiency and reproducibility. Reproducibility, high affinity and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
experiment example 1
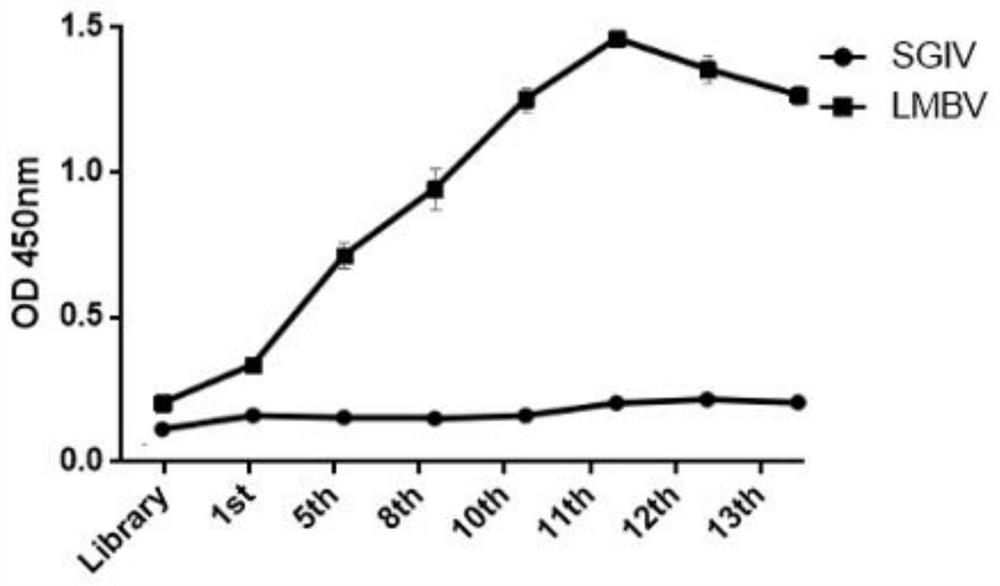
[0085] The purpose of Experimental Example 1 is to measure and analyze the affinity constants of the three nucleic acid aptamers (LA38, LB49, LA13) obtained in the above examples and LMBV respectively. The ELASA method was used to analyze the binding affinity of the nucleic acid aptamer to LMBV, and the specific steps were as follows:
[0086] (1) Dilute the 5'-biotinylated nucleic acid aptamer to different concentrations (0-1000nM), denature at 95°C for 5 minutes, cool on ice for 10 minutes to denature, and then add to the enzyme coated with LMBV Combine in wells of linked plates for 1 hour at 4°C in the dark;
[0087] (2) Wash the corresponding wells with PBS buffer 5 times again, after the incubation, wash 3 times with PBS, add HRP-Strep (1:10000 dilution), and incubate for 30 minutes;
[0088] (3) After washing with PBS for 5 times, use the Pierce TMB substrate kit to develop color, and read the OD corresponding to the well 450 value;
[0089] (4) The mean value of the ...
experiment example 2
[0092] The purpose of Experimental Example 2 is to analyze the toxicity of the three nucleic acid aptamers (LA38, LB49, LA13) obtained in the above examples. Use the kit (CCK-8) to measure the toxicity of the nucleic acid aptamer at the cellular level, and the specific steps are as follows:
[0093] (1) Culture carp epithelial tumor cells (EPC) in a 96-well plate for 24 hours, discard the culture medium, add different concentrations (1-1000nM) of pre-denatured nucleic acid aptamers, and continue to culture at 28°C for 48 hours. Treated EPC cells were used as controls;
[0094] (2) According to the operation method of the kit manual, add 10 μL of CCK-8 solution to each well, and incubate at 28°C for 4 hours;
[0095] (3) Use a microplate reader to detect the absorbance at 450 nm.
[0096] Using the CCK-8 cell viability assay kit, the cell viability results of EPC cells after treatment with different concentrations of nucleic acid aptamers were as follows: Figure 5 As shown,...
experiment example 3
[0105] The purpose of Experimental Example 3 is to analyze the effects of the three nucleic acid aptamers (LA38, LB49, and LA13) obtained in the above-mentioned examples on in vitro resistance to LMBV infection. Specific steps are as follows:
[0106] The random starting library (Library) and the screened 3 nucleic acid aptamers (200nM) were added to the EPC cells respectively as 4 experimental groups, and the EPC cells without any treatment were used as the blank group (Mock for short), After 24 hours, observe the cell growth situation of each group, the results are as follows: Figure 7 As shown, it can be seen that the treatment of EPC cells with nucleic acid aptamers has no effect on cell growth.
[0107] In addition, the random starting library (Library) was incubated with LMBV at 4°C for 1 hour and then added to the EPC cells. As a control group, the three nucleic acid aptamers (200nM) screened were incubated with LMBV at 4°C After 1 hour, it was added to the EPC cells...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com