Universal hybridization enhancer and method for targeted sequencing
A targeted sequencing and enhancer technology, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of unsmooth process, cumbersome operation, unfavorable automation platform establishment, etc., and achieve excellent blocking effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0038] 1. Pre-library construction
[0039] DNA library construction kit (Rapid DNA Lib Prep Kit, ABclonal) and Illumina TruseqDual-index adapter ( Figure 4 ), the pre-library used in this example was constructed on NA12878 gDNA (Coriell) (insert size: ~200bp; number of PCR cycles: 7).
[0040] 2. Hybrid capture
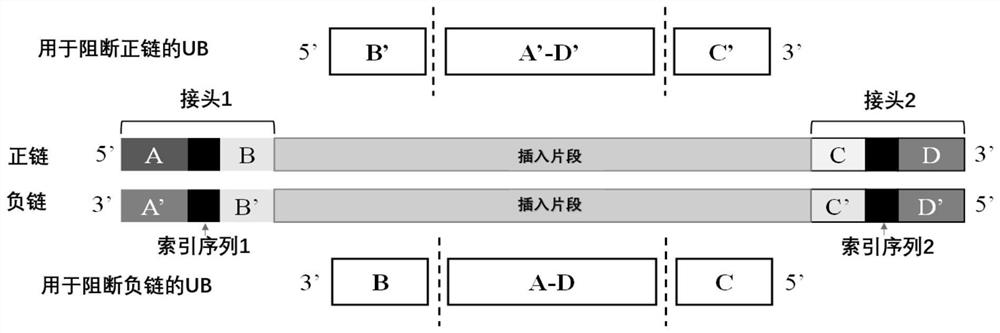
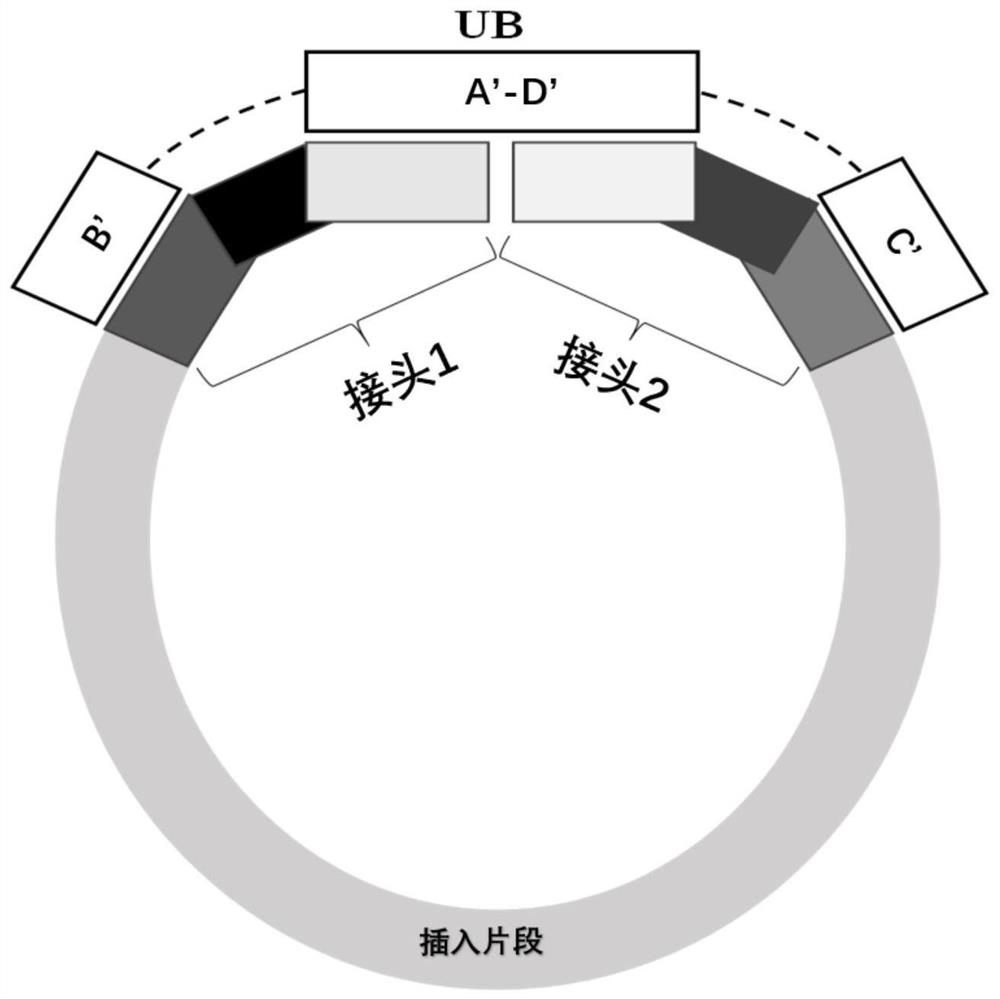
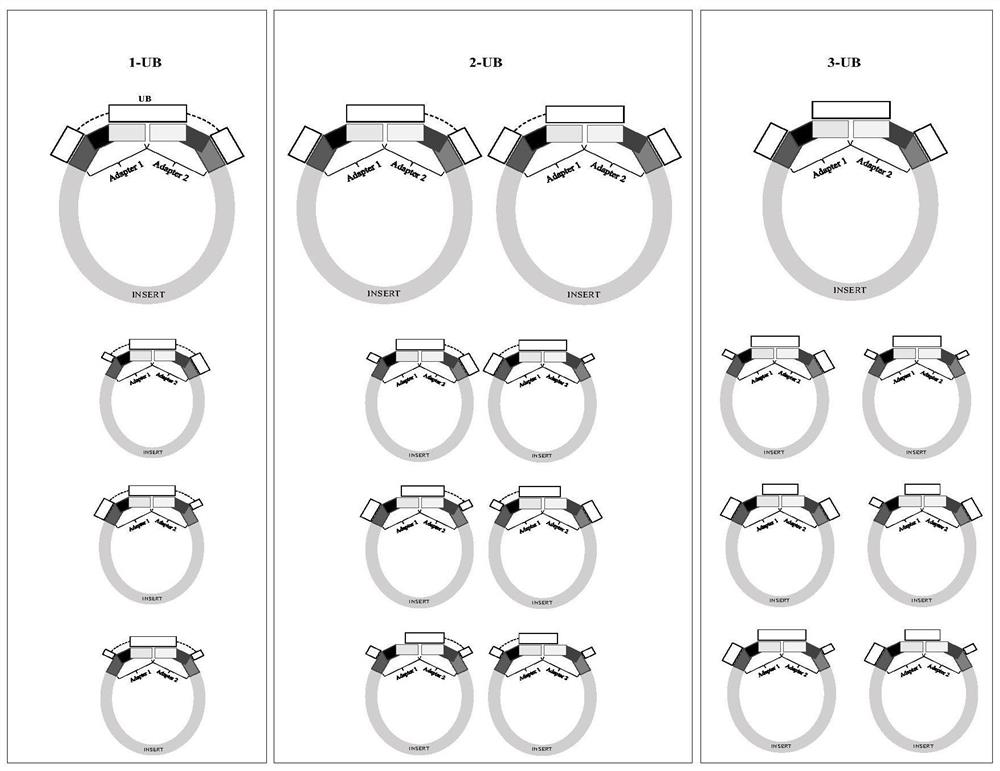
[0041] For B, A-D and C regions, design and synthesize LNA-containing oligonucleotides as components constituting universal hybridization enhancers, wherein oligonucleotides 3-UB 24LNA-B, 3-UB 24LNA-AD, 3- UB 24LNA-C is complementary to regions B, A-D and C, respectively, that is, blocks the above regions. Subsequently, using the specific hybridization enhancer corresponding to the Index sequence one-to-one as a control, hybridization capture was performed on the combination of universal blocking agent components, and the blocking effect was tested. A 4-hour hybridization capture (N=2) was performed following the steps indicated in A-J. The oligonucleotide seque...
Embodiment 2
[0103] This example is used to explore the Tm value of general hybridization enhancers. The method in Example 1 was used to construct the Illumina Truseq Dual-index linker pre-library of NA12878gDNA (Coriell).
[0104] As shown in Table 8, the specific hybridization enhancer and the general hybridization enhancer (1 nmoleEach) containing different amounts of LNA were used to carry out hybridization capture, sequencing and analysis of the pre-library (the same method as in Example 1) (N=2). See Table 1 for the sequence information of hybridization enhancers.
[0105] Table 8
[0106]
[0107] As shown in Table 9, as the Tm value increases, the On-Target ratio of the library gradually increases. When the Tm value of the universal hybridization enhancer reaches 80°C (3-UB 12LNA), its On-Target ratio is significantly improved compared with the On-Target ratio of the sample without the hybridization enhancer; when the Tm value of the universal hybridization enhancer reaches 83...
Embodiment 3
[0111] This example is used to study the blocking effect of universal hybridization enhancers on different Indexes.
[0112] Using the library construction method in Example 1, the pre-library used in this example was constructed on NA12878 gDNA (Coriell). Twelve cases of adapters were constructed with illumina Truseq Dual-index and pre-libraries containing different indexes (insert size: ~200bp; number of PCR cycles: 7). 3-UB 24LNA hybridization enhancer (1 nmole each) was used to carry out hybridization capture, sequencing and analysis of the above 12 pre-libraries (the method is the same as in Example 1).
[0113] As shown in Table 10, 8 kinds of Adapter-1 and 12 kinds of Index,3-UB 24LNA hybridization enhancers of Adapter-2 can obtain stable and efficient blocking effect.
[0114] Table 10
[0115]
[0116]
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting point | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com