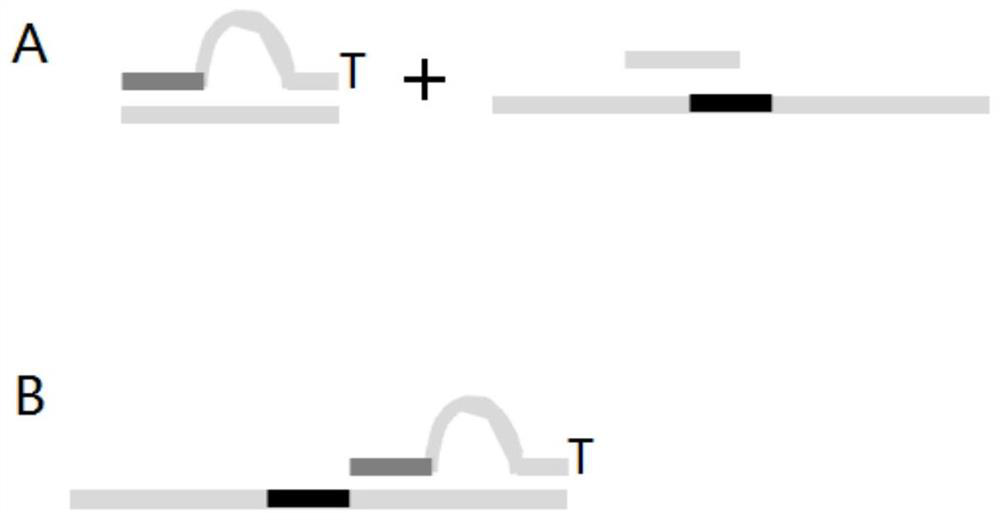
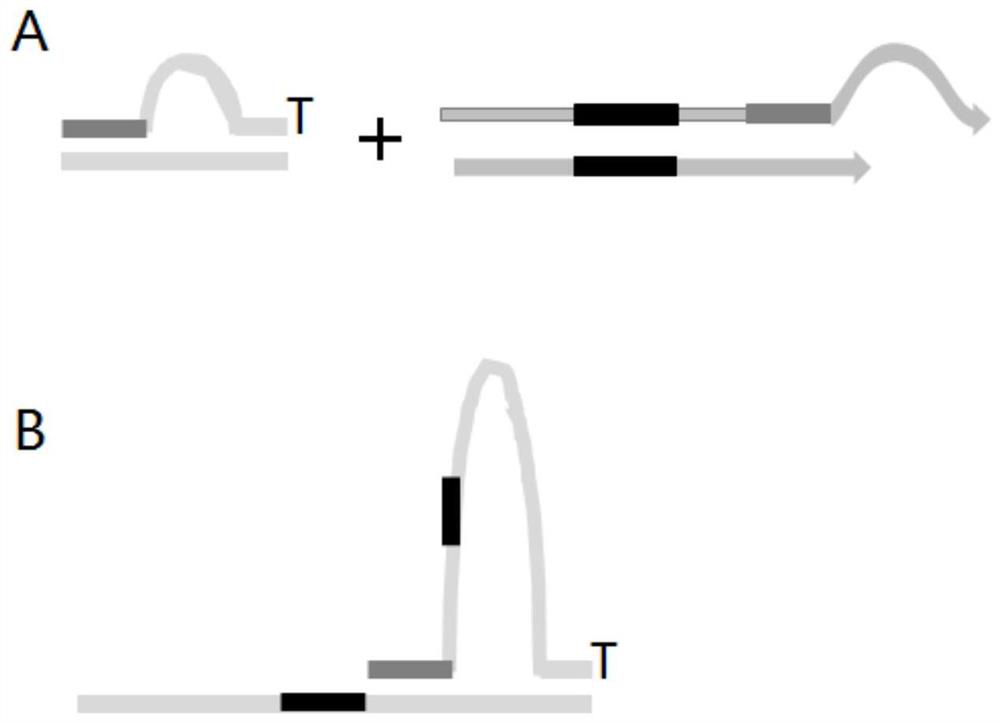
Double-ended library tag composition and application thereof in MGI sequencing platform
A technology of composition and tag group, applied in the field of plasma DNA library construction, which can solve problems such as sample crosstalk
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0081] Example 1 Database construction scheme one and scheme two
[0082] Specific steps: refer to NadPrep TM The only difference in the manual of the DNA library construction kit (for MGI) (201909Version2.0) is the difference in the sequence of the bubble adapter and the sequence of the amplification primer
[0083] (1) Option 1:
[0084] Bubble linker sequence:
[0085] Linker sequence 1 shown in SEQ ID NO:769 and linker sequence 2 shown in SEQ ID NO:770:
[0086] SEQ ID NO: 769: (31bp) / phos / agtcggaggccaagcggtcttaggaagacaa;
[0087] SEQ ID NO: 770 (40bp): ttgtcttcctaacaggaacgacatggctacgatccgact*t.
[0088] Amplification primer 1 shown in SEQ ID NO:771 and amplification primer 2 shown in SEQ ID NO:772:
[0089] SEQ ID NO:771: (64bp)
[0090] / phos / ctctcagtacgtcagcagttnnnnnnnnnncaactccttggctcacagaacgacatggctacga; Wherein, the sequence ( / phos / ctctcagtacgtcagcagtt) before nnnnnnnnnn is recorded as SEQ ID NO:793, and the sequence after nnnnnnnnnn (caactccttggctcacagaac gac...
Embodiment 2
[0120] Example 2 12 sample mixed data split comparisons with 4 balance and 8 balance
[0121] The double-ended labeling scheme can effectively remove the crosstalk between samples (also known as label skipping), but since splitting the data requires correct labels at both ends, the valid sequencing data can be split, so the label balance requirements when using the machine Stricter than single-ended labeling requirements. This application optimizes two sets of schemes of 4 balance and 8 balance. In this embodiment, 4 balance and 8 balance are used respectively, and 12 library mixed samples are tested on the computer to detect the effective resolution rate of each sample by the two sets of schemes. The specific experimental steps and information are as follows:
[0122] Specific steps: refer to NadPrep for the steps of building a library TM Instructions for DNA Library Construction Kit (for MGI) (201909Version2.0), the only difference is that the single-end index adapter is c...
Embodiment 3
[0133] In order to ensure the performance difference between the 8-balanced 48-group tag sequence of this application and the 8-balanced 12-group tag sequence provided by Huada Manufacturing, the 8-balanced 48-group tag sequence of this application was designed in consideration of the 8-balanced 48-group tag sequence provided by Huada Manufacturing. The compatibility of the balanced 12 sets of tag sequences when used on the machine, therefore, there are 3 bases in any two sequences between the 8 balanced 48 sets of tag sequences of this application and the 8 balanced 12 sets of tag sequences provided by Huada Manufacturing base difference.
[0134] Additionally, other major points of difference are:
[0135] 1. The base composition of the tag sequence of the present invention is more balanced, with a GC% content of 40%-60%; while MGI’s GC% content is 20%-80%;
[0136] 2. The tag sequences of the present invention have been calculated for matching with the linker sequences of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com