Polypeptide with binding affinity to chlamydia trachomatis MOMP and application of polypeptide
A Chlamydia trachomatis and affinity technology, applied in the field of biomedicine, can solve the problems of high cost, affecting the wide application of monoclonal antibodies, and large toxic and side effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0109] Example 1. Library Construction and Screening Research of Ct MOMP Binding Polypeptides
[0110] A random combinatorial library of phage-displayed Ct MOMP-binding polypeptides was constructed, that is, a library of many different SPA domain-related polypeptides, and Ct MOMP-binding polypeptides were screened from the library and their affinities were identified.
[0111] 1. Construction and identification of a random combinatorial phage display library of Ct MOMP-binding peptides
[0112] According to the amino acid sequence and structure of wild-type SPA-Z (Nilsson B et al., Protein Eng.1987; 1 (2): 107-113), random primers were designed for the coding sequences corresponding to its three helical regions, and amplified by PCR. A SPA coding sequence that can lead to random amino acid mutations was added and named SPA-N. According to the conventional method of molecular cloning, the SPA-N coding sequence was cloned into the pCANTAB5E vector through the SfiI and NotI site...
Embodiment 2
[0122] Example 2, Ct MOMP-binding polypeptide recombinant plasmid construction and prokaryotic protein expression and purification
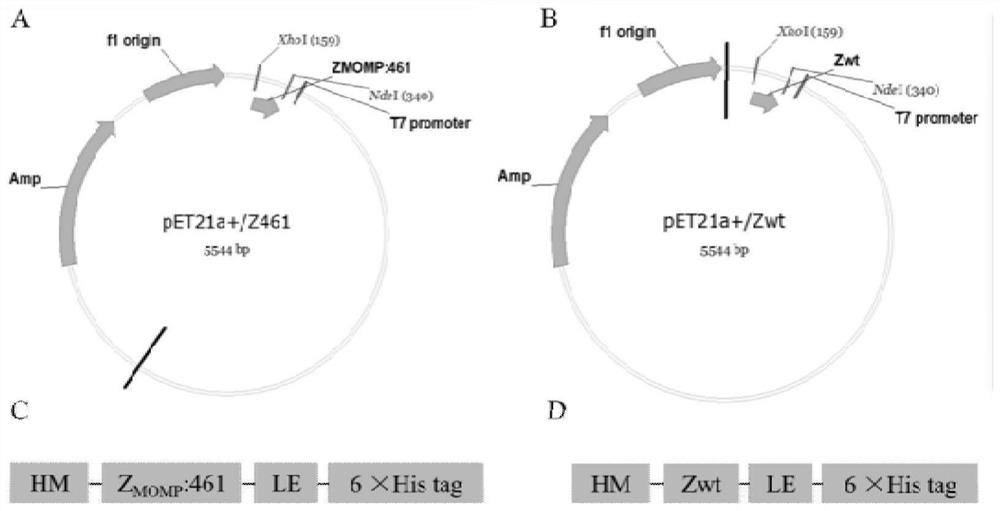
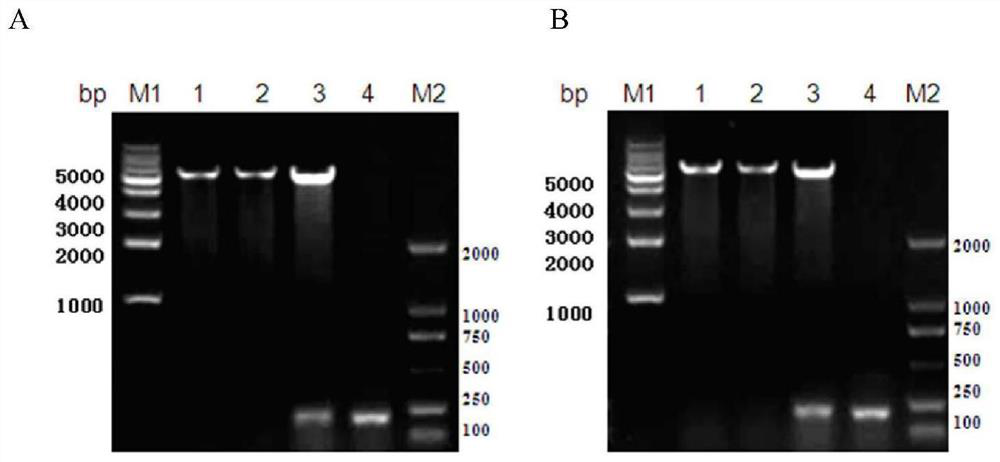
[0123] 1 clone with a higher ELISA read was selected as before ( figure 1 Z in Ct MOMP), and Zwt as a negative control for Ct MOMP-binding polypeptides. In order to perform functional testing on the screened affibody molecules, construct recombinant plasmids, express and identify prokaryotic proteins, and prepare purified proteins.
[0124] 1. Construction and identification of recombinant plasmid of pET21a(+) / affibody
[0125] Design PCR primers and upstream primers with reference to the affibody gene sequence (GenBank: GY324633.1)
[0126] 5'GGGAATTC CATATG GTTGACAACAAATTCAACAAAGAA 3' (SEQ ID NO: 6, italics and underline indicate Ned I restriction site), downstream primer 5'CCG GAATTC CGTTTCGGAGCCTGAGCGT 3' (SEQ ID NO: 7, italics and underline indicate the XhoI restriction site); the correct sequencing of the screened tertiary library mo...
Embodiment 3
[0129] Example 3, Z Ct MOMP Binding to Ct MOMP protein
[0130] To identify Z Ct MOMP Binding specificity to Ct MOMP, using surface plasmon resonance (SPR) analysis of screened Z Ct MOMP The binding affinity and specificity of its control Zwt affibody to the target protein MOMP.
[0131] 1. Preparation and identification of Ct MOMP
[0132] The pET21a(+) / Ct MOMP constructed and stored in the laboratory was transformed into Escherichia coli BL21(DE3), and the recombinant protein was expressed after being induced by IPTG. The protein was purified by Ni-NTA affinity chromatography, and the Japanese white ear was routinely immunized Serum antibodies were prepared from rabbits. As a result, SDS-PAGE electrophoresis showed that an obvious protein band appeared at a relative molecular mass (Mr) of about 40kDa, which was consistent with the expected protein Mr size ( Figure 5 A); using the mouse anti-6×His mAb as the primary antibody for Western blot analysis, it can be seen tha...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com