Model for predicting cell proliferation activity by taking 87 genes as biomarkers
A biomarker and cell proliferation technology, applied in the field of predicting cell proliferation activity, can solve problems such as differences in living environment, inability of cells to be cultured in vitro, proliferation ability cannot reflect proliferation ability, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
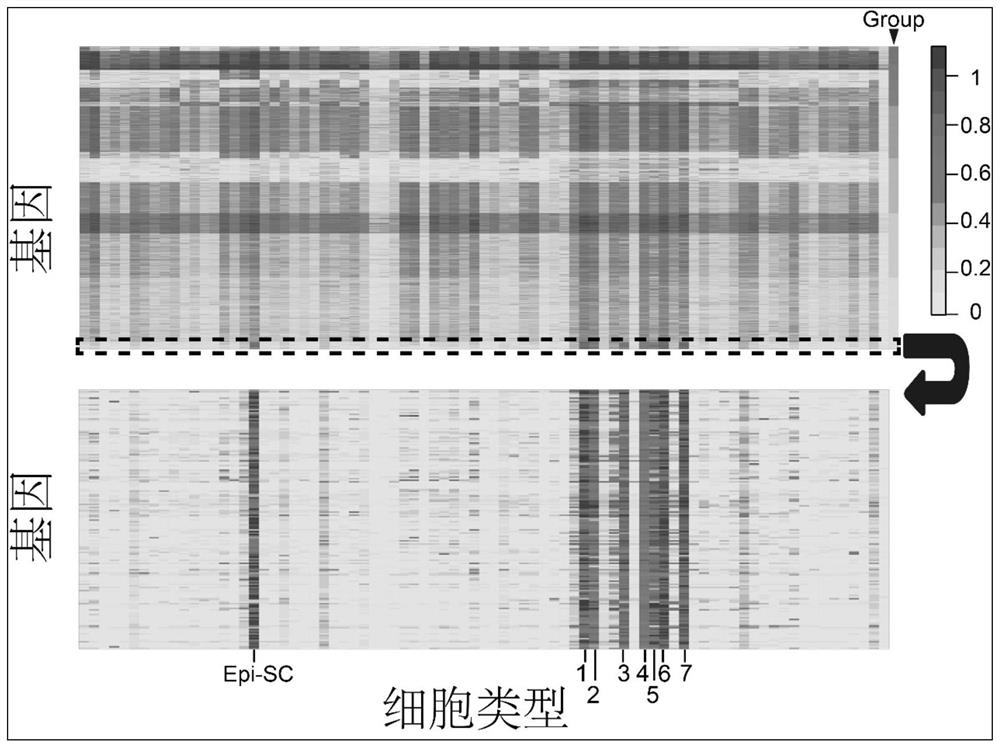
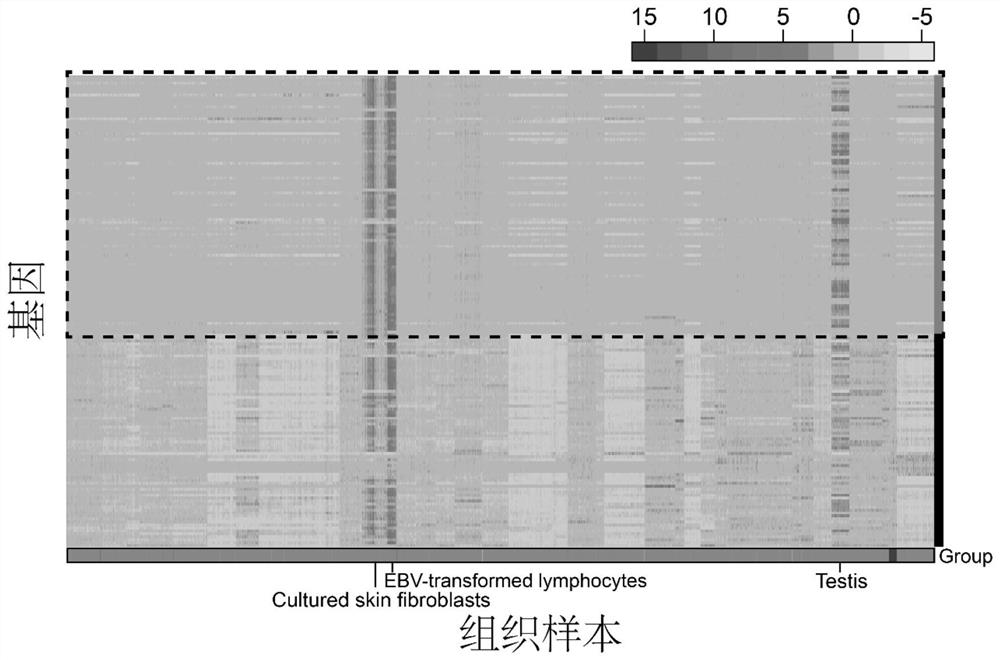
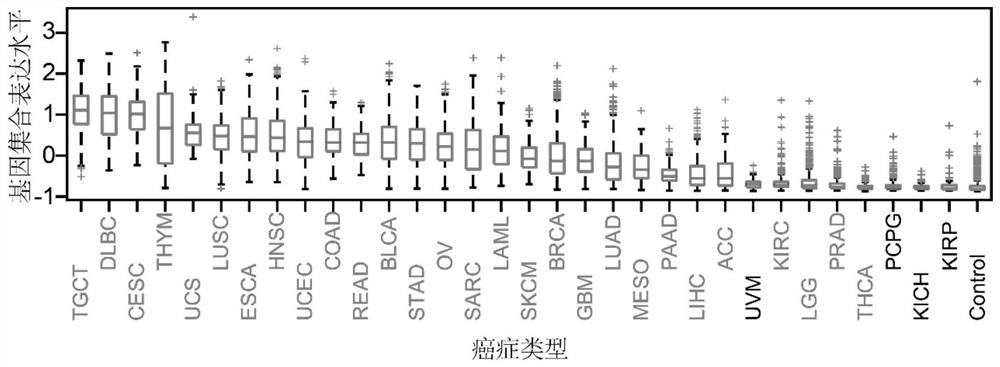
[0036] Example 1: Use the Tabula Muris database, TCGA database, GTEx database and CCLE database to establish a cell proliferation gene set containing 87 genes, predict the proliferation activity of 81 different normal cell types collected in the Tabula Muris database, and assist in judging TCGA Whether there are a large number of normal cells with proliferation activity in the cancer tissues in the database guides cancer treatment and evaluation methods for cell proliferation markers.
[0037] (1) Data collection
[0038] 53,760 single-cell RNA-Seq data from 81 different normal cell types generated by Smart-Seq2 single-cell sequencing technology were obtained from the Tabula Muris database. The RNA-Seq data of 9630 carcinomas and paracancerous tissues of 32 cancers were obtained from The Cancer Genome Atlas (TCGA) database, and the prognosis data of 31 cancers were also obtained. RNA-Seq data for 17,382 tissues of 54 tissues were obtained from the GTEx database. The RNA-Seq ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com