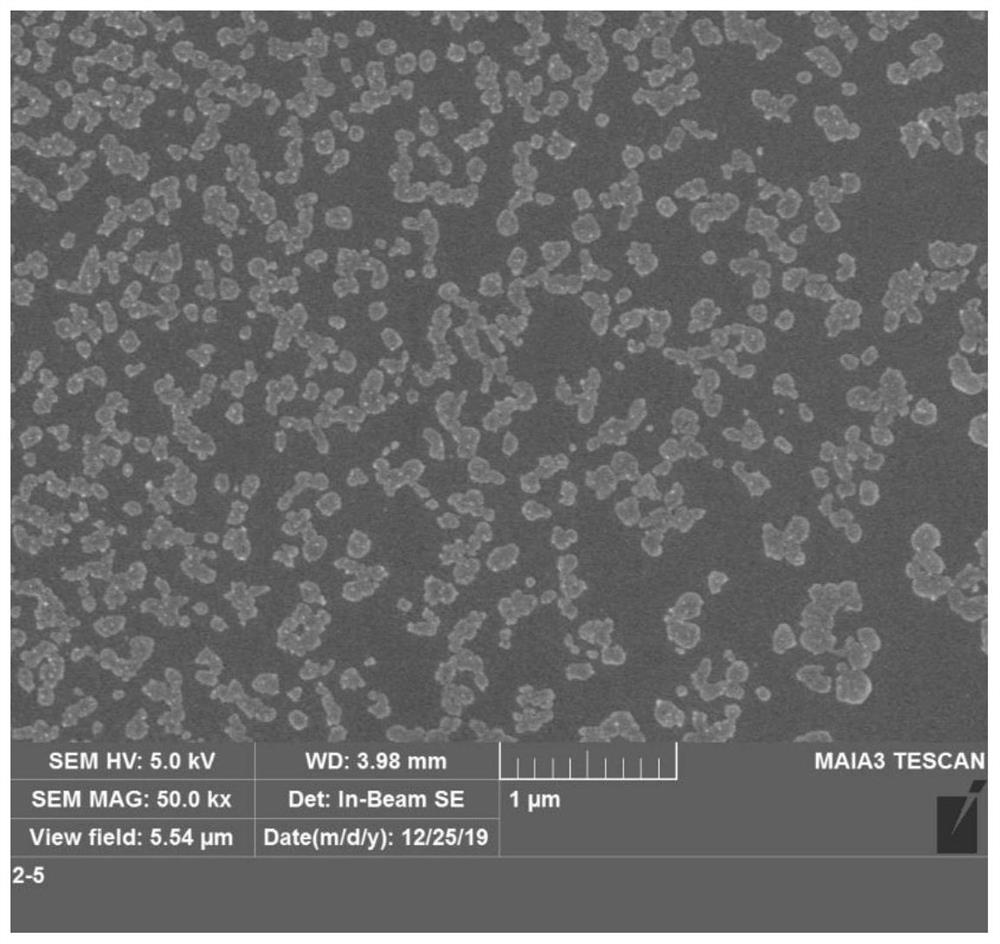
Self-assembled nano material as well as preparation method and application thereof
A nanomaterial, self-assembly technology, applied in the field of biomimetic catalytic materials, can solve the problems of variability, protein structure unfolding, active structure destruction, etc., and achieve the effect of low price
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] (1) Preparation of self-assembly mimetic enzymes
[0028] To 96 μL 50 mM Na 2 HPO 4 / NaH 2 PO 4 Add 1 μL of 100 μM G-DNA, 2 μL of 1 mg / mL Fmoxy-lysine and 1 μL of 1000 μM CuCl to the buffer solution (pH 7.0) 2 , samples were added at intervals of 10 s, followed by mixing at 25 °C for 30 min. Wherein, the sequence of G-DNA is GGGTAGGGCGGGCGGG (SEQ ID NO.1), which is purified by HPLC (high performance liquid chromatography) or PAGE (polyacrylamide gel electrophoresis).
[0029] (2) Catalytic activity test
[0030] Add 1 μL of 100 mM tetramethylbenzidine (dissolved in DMSO) to 100 μL of simulated enzyme solution sequentially, the interval between sample additions is 5 s, and record the absorbance at 652 nm (the oxidation intermediate product of tetramethylbenzidine) over time. Change, through the molar extinction coefficient of the oxidation product of tetramethylbenzidine (39000M cm -1 ), calculate the oxidation rate of tetramethylbenzidine. The switching number o...
Embodiment 2
[0032] To 90 μL 50 mM K 2 HPO 4 / KH 2 PO 4 Add 1 μL of 1000 μM G-DNA, 8 μL of 200 mM peptide and 1 μL of 500 μM CuCl to the buffer solution (pH 7.0) 2 , samples were added at intervals of 10 s, followed by mixing at 25 °C for 30 min. Wherein, the sequence of G-DNA is GGTAGGCGGCGGTGGCGGCGGAGG (SEQ ID NO.2), purified by HPLC or PAGE; the sequence of polypeptide is phenylalanine-lysine-lysine, purified by HPLC.
[0033] (2) Catalytic activity test
[0034] Add 1 μL of 100 mM phenol (dissolved in DMSO) and 1 μL of 200 mM 4-aminoantipyrine solution to 100 μL of simulated enzyme solution in sequence. Pyrine dye) with time, through the molar extinction coefficient of indoxyl amino antipyrine dye, calculate the oxidation rate of phenol. The switching number on the copper ion surface is 0.65s -1 , the catalytic oxidation rate is 7.85mM -1 the s -1 .
Embodiment 3
[0036] (1) To 90 μL of 50 mM MES / MES sodium salt buffer solution (pH 7.0, containing 50 mM NaCl), sequentially add 5 μL of 200 mM guanosine-5′-monophosphate disodium salt, 4 μL of 2 mM polypeptide and 1 μL of 1000 μM CuCl 2 , samples were added at intervals of 10 s, followed by mixing at 20 °C for 30 min. Wherein, the sequence of the polypeptide is phenylalanine-lysine-phenylalanine-lysine, and is purified by HPLC.
[0037] (2) Catalytic activity test
[0038] Add 1 μL of 100 mM 2,4-dichlorophenol (dissolved in DMSO) and 1 μL of 200 mM 4-aminoantipyrine solution to 100 μL of simulated enzyme solution in sequence. Indoxyl amino antipyrine dye) changes with time, through the molar extinction coefficient of indoxyl amino antipyrine dye, calculate the oxidation rate of 2,4-dichlorophenol. The switching number on the copper ion surface is 0.22s -1 , the catalytic oxidation rate is 1.64mM -1 the s -1 .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com