Application of mir-29a gene in detecting liver cancer and liver fibrosis, and construction method of gene conditional knock-in mice
A mir-29a, 1.mir-29a technology, applied in biochemical equipment and methods, other methods of inserting foreign genetic materials, genetic engineering, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
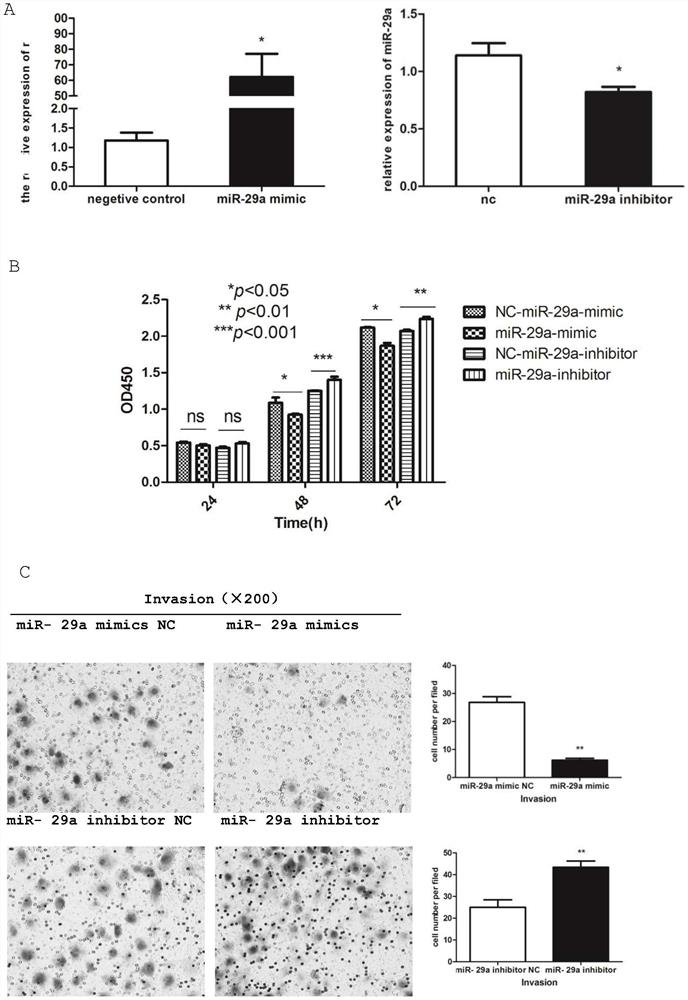
[0041] Example 1 Analysis of the expression of mir-29a in human liver cancer tissues and liver cancer cell lines and its effect on liver cancer cells
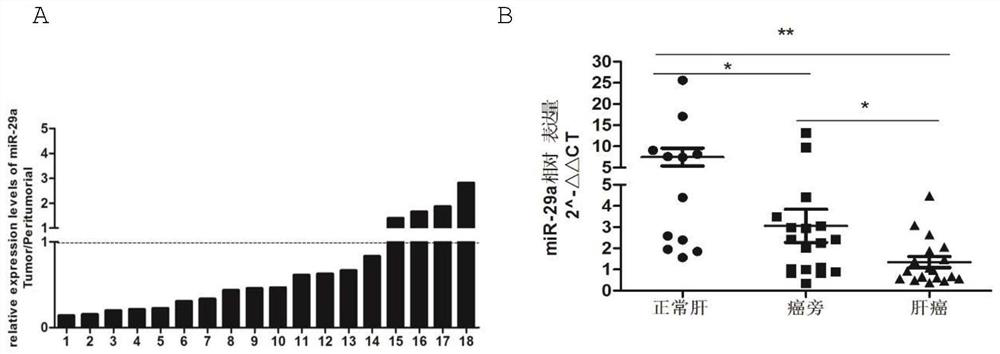
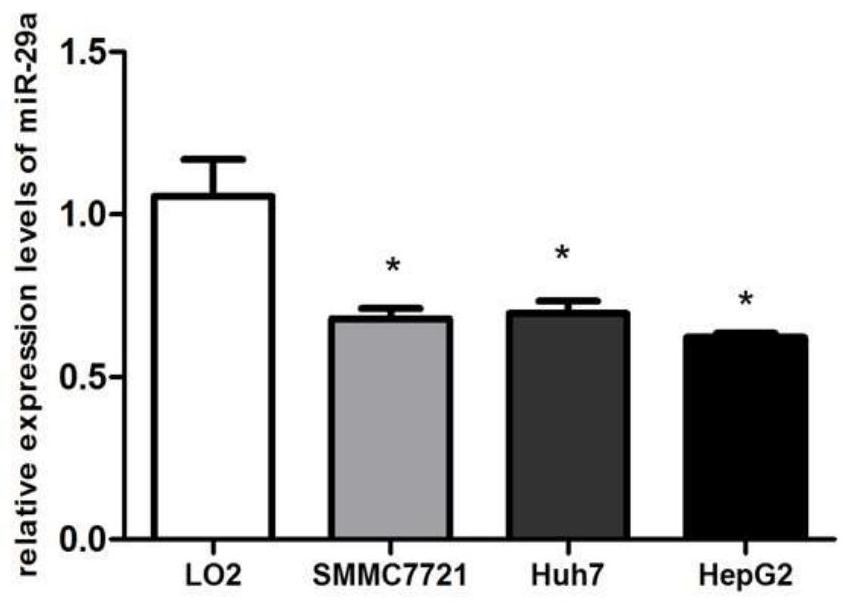
[0042] 1. Detection of miR-29a expression level in human normal liver tissue / hepatocellular carcinoma / paracancerous tissue specimens and liver cancer / normal liver cell lines
[0043] Human liver samples were collected and divided into normal liver tissue (n=12), liver cancer group (n=18) and paracancerous group (n=18). RT-PCR method was used to detect the expression level of miR-29a in human normal liver tissue, liver cancer and paracancerous tissue samples, and GraphPadPrism 5 software was used to analyze and count whether there were significant differences in the relative expression levels among the four groups; the liver cell lines were cultured under the above conditions (L02, HepG2, Huh7, SMMC7721), RT-PCR method was used to detect the expression level of miR-29a in normal liver cells LO2, liver cancer cell lines HepG2, Hu...
Embodiment 2
[0058] Example 2 Effect of mir-29a acting on hepatic stellate cells on liver fibrosis in Schistosoma japonicum mice
[0059] 1. Establishment of liver fibrosis model of Schistosoma japonicum mice
[0060] C57BL / 6 mice (6-8 weeks), 10 in each group were randomly divided into experimental groups (infection 4W, 6W, 8W, 12W) and control groups (infection 0W), the infection operation steps were as follows: (1) The snails with cercariae were placed under light, water temperature water, The cercariae can be seen escaping and floating on the water surface. (2) Pick a little water containing cercariae from the front end of the inoculation loop and put it on the cover glass, and count (16±1) under the dissecting microscope. (3) Remove 1×1cm of mouse abdomen 2 Rat hair, attach the slide to the abdomen for 15-20 minutes. The control group was similarly treated with sterile saline. (4) Raised in a P3 animal experiment center.
[0061] 2. Pathological staining of liver tissue of mo...
Embodiment 3
[0070] Example 3 Preparation and application of mir-29a gene conditional knock-in mice (H11-MIR29A-cKI mice)
[0071] 1. Preparation of mir-29a gene conditional knock-in mice (H11-MIR29A-cKI mice)
[0072] 1. Construction of human MIR29A-H11-cKI donor vector
[0073] 1) Synthesize the mir-29a gene sequence (NCBI Reference Sequence: NR_029503.1), as shown in SEQ No.1;
[0074] 2) Acquisition of the connecting skeleton: construct the basic skeleton of the H11 site (the Hipp11 (H11) locus is located in the intergenic region between the Eif4enif1 and Drg1 genes on mouse chromosome 11), and name it VB027; use the AsiSI / PacI double enzyme Cut the VB027 vector backbone, and recover the 11.5kb fragment by gel;
[0075] 3) Acquisition of the ligated fragments: use EcoRI to digest the Mir-29 gene synthesis product to synthesize a plasmid, and recover the fragments from the gel;
[0076] 4) Use In-fusion ligase to connect the fragment + backbone according to the operation guide of NEB...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com