Preparation method and detection method of DNA specific methylation site
A detection method, a methylation technology, applied in the field of nanobiology and detection analysis, can solve the problems of false positives, high sample requirements, low specificity, etc., and achieve direct visual detection, good biocompatibility, and realization The effect of pleochroism
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
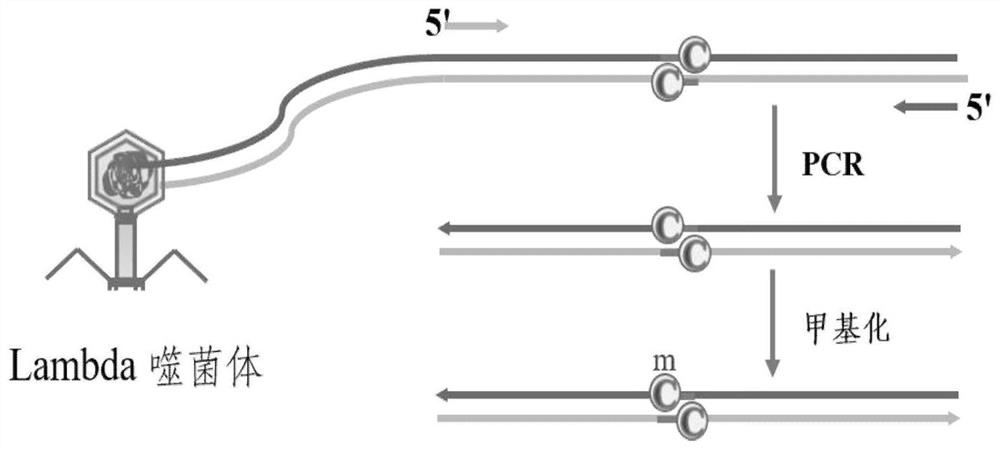
[0033] Such as figure 2 As shown, it is a schematic diagram of the design and preparation of Lambda DNA-specific methylation sites for the embodiment of the present invention, and its specific operations are:
[0034] (1) Preparation of DNA fragments. Select the GpC site to be methylated from the entire Lambda DNA; design the upstream primer and downstream primer so that the GpC site is placed between the upstream primer 1 and the downstream primer 1; set up the PCR reaction program, and use LongAmpTaq DNA polymerization Enzymes for PCR reactions. The sequences of the upstream and downstream primers during the PCR reaction were designed by the software Primer-BLAST. The 50 μL reaction system contains: 10 μL 5×LongAmp Taq reaction solution, 2 μL dNTP mixture (2.5 mM), 2 μL LongAmp Taq DNA polymerase, 2 μL upstream primer 1 (10 μM), 2 μL downstream primer 1 (10 μM ), 0.5 μL LambdaDNA (2 ng / μL), 31.5 μL ultrapure water. The components were mixed quickly on ice and transferre...
Embodiment 2
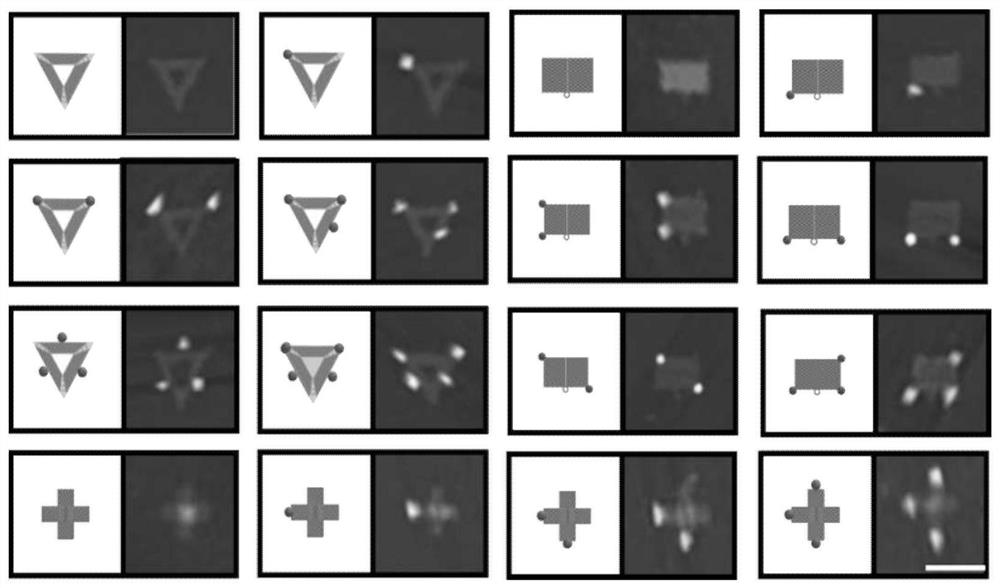
[0040] Such as figure 1 Shown is a schematic diagram of the design of 16 kinds of DNA origami probes in the embodiments of the present invention. Regarding the design and preparation of the 16 kinds of DNA origami probes in the embodiments of the present invention, the specific operations are as follows:
[0041](1) Preparation and purification of basic DNA origami probes: 2.5 μL M13mp18 DNA, 5 μL unmodified staple strand (500 nM), 5 μL staple strand with capture strand at the end (500 nM), 5 μL end with The staple chain (500 nM) of the extended chain, 10 μL of 10×TAE buffer (Mg2+ concentration 125 mM), and 72.5 μL of ultrapure water were evenly mixed so that the final concentration of the backbone chain and the short chain was 1:10:10 : molar ratio of 10; the mixture was placed in a PCR instrument, annealed from 95°C to 25°C at a rate of 0.1°C / 10s, and the product was collected; the product was purified and recovered using Amicon Ultra centrifugal Filters 100K ultrafiltration...
Embodiment 3
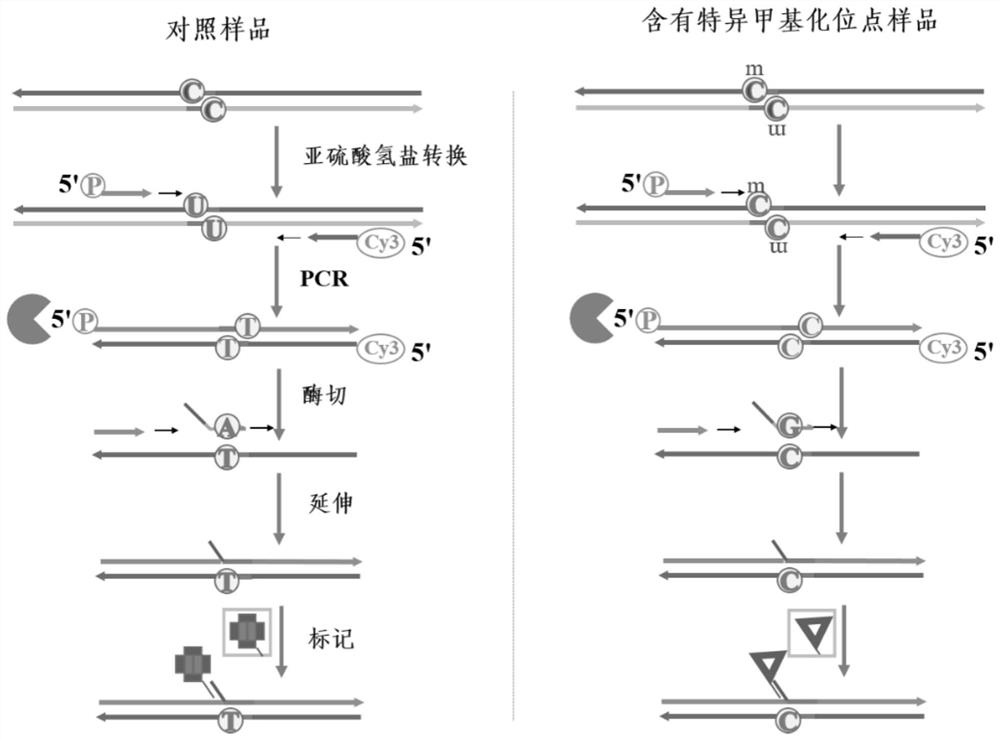
[0046] Such as image 3 Shown is a schematic diagram of the detection of DNA-specific methylation site samples and control samples in the embodiment of the present invention, Figure 4 It is the effect diagram of the DNA origami probe of the embodiment of the present invention respectively marking the two groups of Lambda DNA control samples containing specific methylation sites and not containing specific methylation sites, about the DNA specific methylation of the present invention The corresponding sites in the chemical sites and the control samples are detected, and the specific operations are as follows:
[0047] (1) Bisulfite treatment: Weigh 1.88 g of sodium bisulfite, add ultrapure water to dilute, and titrate the solution with NaOH solution (3 M) to pH 5.0, the final volume is 5 mL, and configure as sodium bisulfite The solution is ready for use; take 50 μL of the product of step 2 in a 1.5 mL centrifuge tube, add 5.5 μL of newly prepared NaOH solution (3 M), and bat...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Full length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com