A method for metabolically engineering Escherichia coli to prepare α-ketoisovaleric acid
A recombinant Escherichia coli, heterogeneous technology, applied in the field of bioengineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
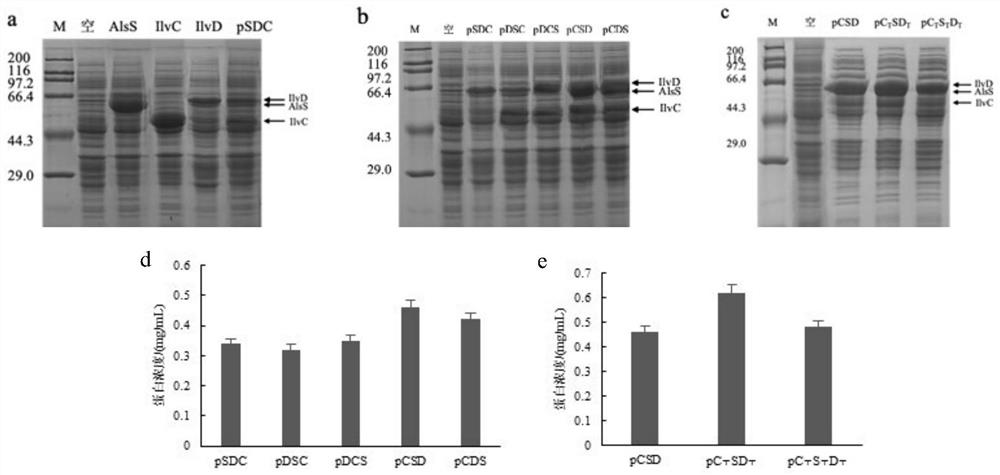
[0034] Example 1: Construction and optimization of acetyl lactate synthetase, an acetyl lactate isomeric reductase and dihydroxy acid dehydrat
[0035] 1) Amino acid sequence of acetyl lactic acid synthetase, such as SEQ ID NO.1, nucleotide sequence, such as SEQ ID NO.2; amino acid sequence of an acetyl lactate isomorbed, as shown in SEQ ID NO.3, The prime sequence is shown in SEQ ID NO.4. The amino acid sequence of dihydroxy acid dehydratase, as shown in SEQ ID NO.5, the nucleotide sequence such as SEQ ID NO.6, according to the purpose gene and the carrier, and is designed to design the primer (such as Table 1);
[0036] Table 1 Plasmid construct primer design
[0037]
[0038]
[0039] Note: Underline marking is an enzyme cleaning point
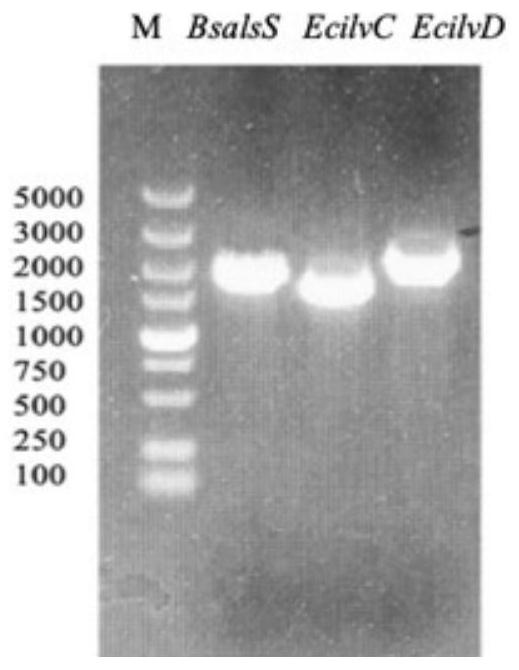
[0040] 2) The E.COLI MG1655 genome is a template, respectively, and cloned the gene BSALSS, ECILVC, ECILVD. The plasmid pETDUET-1 with cloned gene BSALSS, ECILVC, and ECILVD were double-cutted by the obtained enzyme digestion in Table 1, an...
Embodiment 2
[0053] Example 2: Method for the construction of alpha-ketoisukoic acid engineering strains of the knockout competitive metabolic pathway
[0054] 1) Departure strain is the main coli B0016-050 (ΔAck-PTA, ΔPFLB, ΔAck-PTA, ΔPFLB, ΔAdhe, ΔFrda, ΔLDHA) (disclosed in literature z et al. " Appl.biochem. Biotechnol., 2016,178: 324-37. In addition to the coding genes of by-product acetic acid, formic acid, ethanol, succinic acid, lactic acid synthesis, can avoid a lot of these metabolic by-products Synthesis, and provide sufficient pyruvate precursors for fermentation of α-ketoisuate. In order to ensure the normal exercise of the T7 promoter, the T7 RNA polymerase (T7 RNAP) gene is integrated on the genome, and also to block the further decomposition of α-keto ketoikoic acid, encoding the chromosome upper strand amino acid amino enzyme encoding gene ILVE Using the RED recombination method for knockout.
[0055] 2) The genome is a template in Escherichia coli E.COLI BL21 (DE3), and the P3...
Embodiment 3
[0076] Example 3: Construction Method for Coenzyme NADPH Circulating Regenerated α-ketoisuate
[0077] 1) Template is a template in Escherichia coli genomes and plasmid Pacycduet, P49 + P50, P51 + P52 in Table 3, and amplified PNTab fragment and plasmid skeleton PACYC, and the two will be subjected to Gibson assembled to obtain recombinant plasmid PACYC-PNTAB, respectively. .
[0078] 2) The plasmid pACYC-PNTAB and PKD13 are templates, P53 + P54, P55 + P56 in Table 3, and amplified to obtain the plasmid skeleton PACYC-PNTAB and KAN fragments, and the two will be subjected to Gibson assembly to obtain recombinant plasmid PACYC. -kan-pntab.
[0079] 3) Template with plasmid pacyc-kan-pntab, P57 + P58, P59 + P60, P61 + P62 in Table 3, to perform full-quality PCR, to obtain a T7 promoter of different strength, and delete the T7 promoter Laco gene to get recombinant plasmid PACYC-KAN-T7 100 PACYC-KAN-T7 92 PACYC-KAN-T7 16 .
[0080] 4) Particles PACYC-KAN-T7, respectively 100 PACYC-KAN...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com