Method for detecting RNA m6A with high flux, high sensitivity and single base resolution and application thereof
A high-sensitivity, high-throughput technology, applied in the field of high-throughput and high-sensitivity single-base resolution detection of RNAm6A, can solve the problems of cumbersome miCLIP process, difficult to achieve m6A detection, and high experimental conditions, and achieve antibody enrichment efficiency. The effect of improving, improving sensitivity, and reducing false positive rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
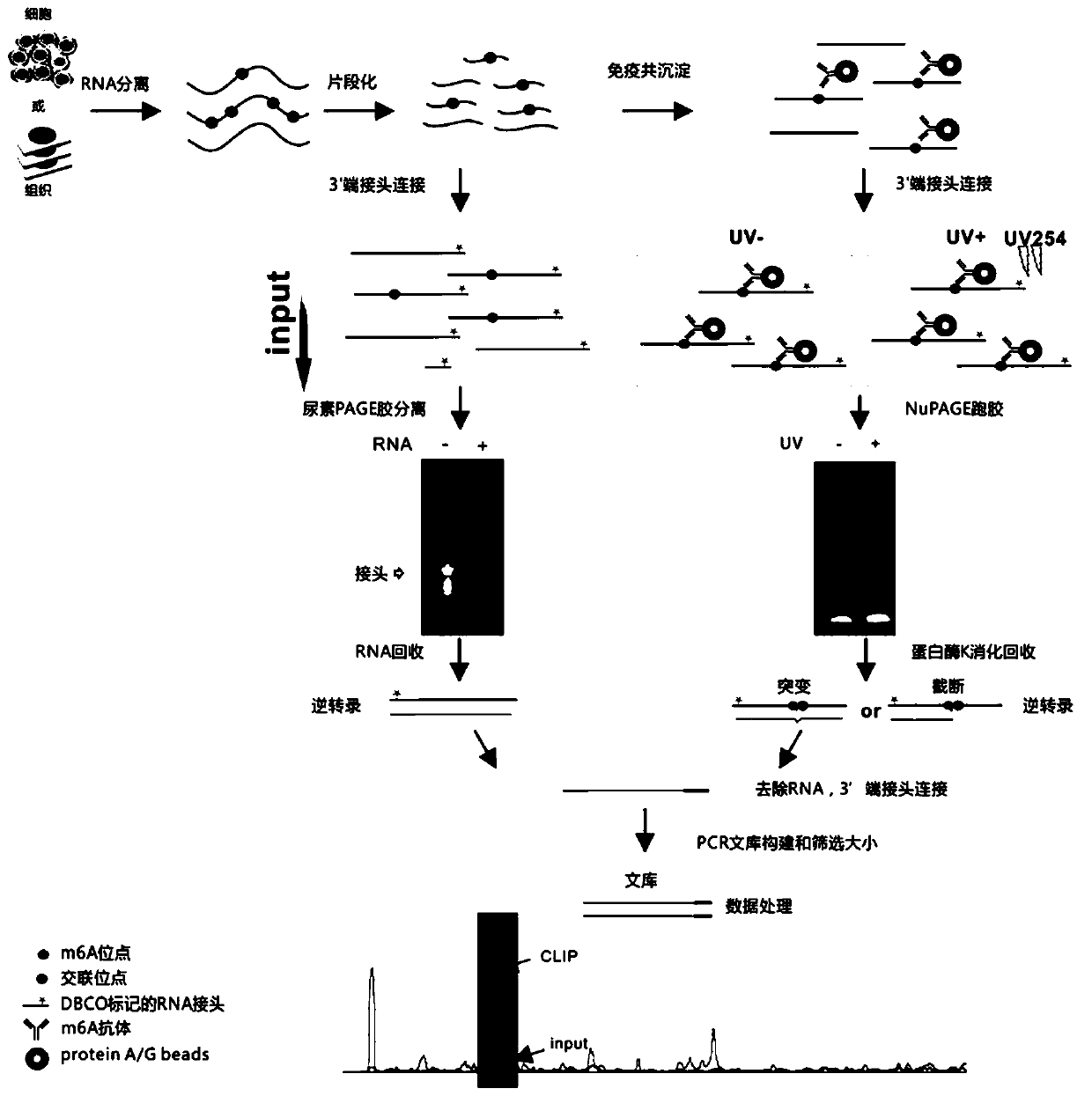
[0072] The application of hmCLIP in fine-needle aspiration paraffin specimens of thyroid cancer involves the following steps in sequence (flow chart as shown in figure 1 shown):
[0073] 1. Obtain total RNA
[0074] Three to five fine-needle aspiration paraffin specimens of thyroid cancer were taken, and total RNA was extracted with QIAGEN RNeasy FFPE kit. The specific method is as follows:
[0075] 1) Dewaxing of paraffin sections: take 3 to 5 paraffin sections, depending on the size of the section tissue, put them in a 1.5ml nuclease-free EP tube, add 1ml xylene, vortex fully for 10s, and centrifuge at the maximum speed for 2min;
[0076] 2) Remove the supernatant as much as possible, add 1ml of absolute ethanol to the precipitate, vortex to mix, and centrifuge at the maximum speed for 2min;
[0077] 3) Remove the supernatant as much as possible, open the lid and place at room temperature for 10 minutes until the residual ethanol evaporates;
[0078] 4) Add 150ul PKD buff...
Embodiment 2
[0269] Referring to steps 1 to 5 in Example 1, the difference is that the cross-linking energy and time of RNA and m6A antibody in step 4 (co-immunoprecipitation and cross-linking) are different, and are divided into the following 6 groups of experiments:
[0270] (1) No UV crosslinking (named UV-);
[0271] (2) Use Spectroline UV 254nm Instrument (model: XLE-1000) for cross-linking, cross-linking energy: 150mj / cm 2, Number of cross-links: 2 times (named: 150mj / cm 2 *2);
[0272] (3) Use Spectroline UV 254nm Instrument (model: XLE-1000) for cross-linking, cross-linking energy: 150mj / cm 2 , Number of crosslinks: 4 times (named: 150mj / cm 2 *4);
[0273] (4) Use Spectroline UV 254nm Instrument (model: XLE-1000) for cross-linking, cross-linking energy: 300mj / cm 2 , Number of cross-links: 2 times (named: 300mj / cm 2 *2);
[0274] (5) Use Spectroline UV 254nm Instrument (model: XLE-1000) for cross-linking, cross-linking energy: 400mj / cm 2 , Number of crosslinks: 2 times (...
Embodiment 3
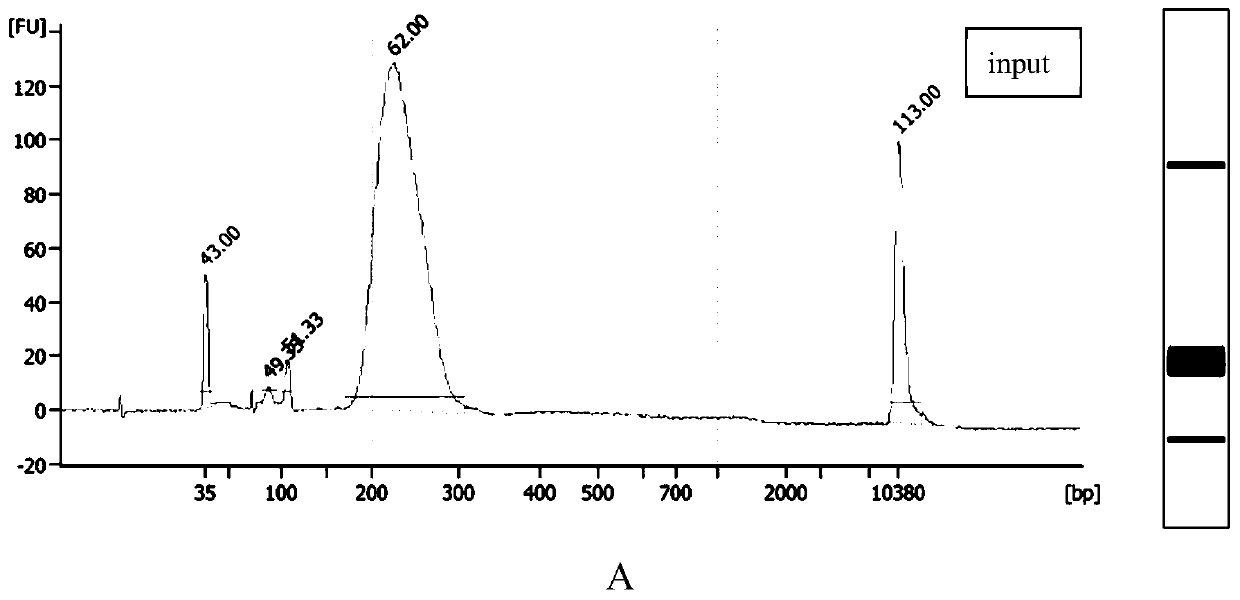
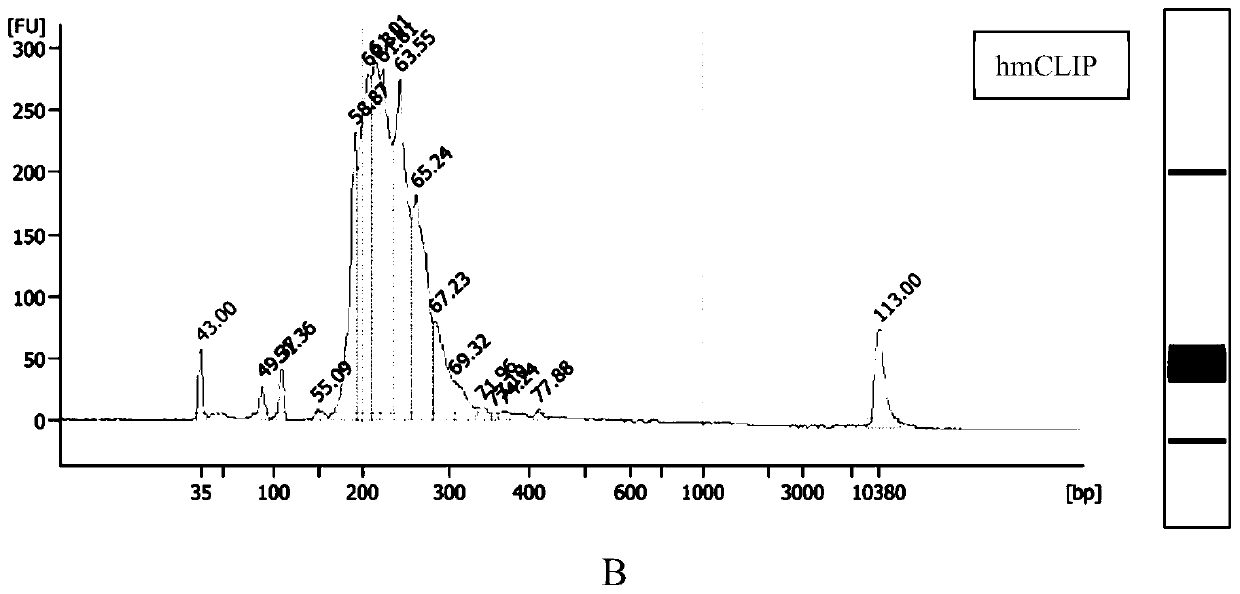
[0278] The method is the same as that of Example 1 (steps 1 to 5.9), the difference is: the concentration of RNA Ligase High conc when the RNA adapter is connected in step 5, the 3' end is connected to the RNA adapter in step 6, and the 3' adapter of cDNA in step 7 is built. For high concentration ligase (T4 RNA Ligase 1, High Concentration (NEB: M0437M)) and common ligase (T4 RNA Ligase, NEB: M0204S) (initial concentrations are 30,000U / ml and 10,000U / ml respectively), Its ligation efficiency as well as the reproducibility of hmCLIP such as Figure 8 Shown (reproducibility of hmCLIP is a repeat of Examples 1 to 5.9)).
[0279] From Figure 8 It can be seen from the figure that the signal of the ligation product obtained by using a high concentration of ligase to connect the fluorescently-labeled adapter is significantly higher than that of the common ligase, and the reproducibility is also high.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com