Nucleic acid sequence detection method based on QCM
A detection method and nucleic acid sequence technology, applied in the field of nucleic acid sequence detection based on QCM, can solve problems such as dependence on enzyme activity, complicated steps, and long time consumption, and achieve high specificity, promote in-situ growth, and inhibit non-specific crystallization Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1 A QCM-based nucleic acid sequence detection method
[0045] (1) Cleaning of the wafer: Put the wafer (the quartz crystal microbalance sensor wafer is produced by Biolin Scientific in Sweden, its diameter is 14mm, the basic frequency is 5MHz) in piranha washing liquid (H 2 SO 4 : H 2 O 2 =3:1 volume ratio) soak for 10 minutes, then follow H 2 O: H 2 O 2 : NH 3 ·H 2 The lye is prepared with a volume ratio of O=5:1:1, the prepared lye is heated and boiled on an electric heating mantle, and the wafer is immersed in the boiling lye for 15 minutes; keep the lye in a boiling state during the soaking process. Rinse with ultrapure water and soak in ultrapure water for 5 minutes, then rinse with absolute ethanol and soak in absolute ethanol for 5 minutes; blow dry with nitrogen, and place the dried wafer in a clean bench under ultraviolet light for 20 Minutes to remove organic matter on the surface;
[0046] (2) Modification of the chip:
[0047] ① Spread the cleaned wafer fl...
Embodiment 2
[0065] Example 2 Verification of signal amplification ability
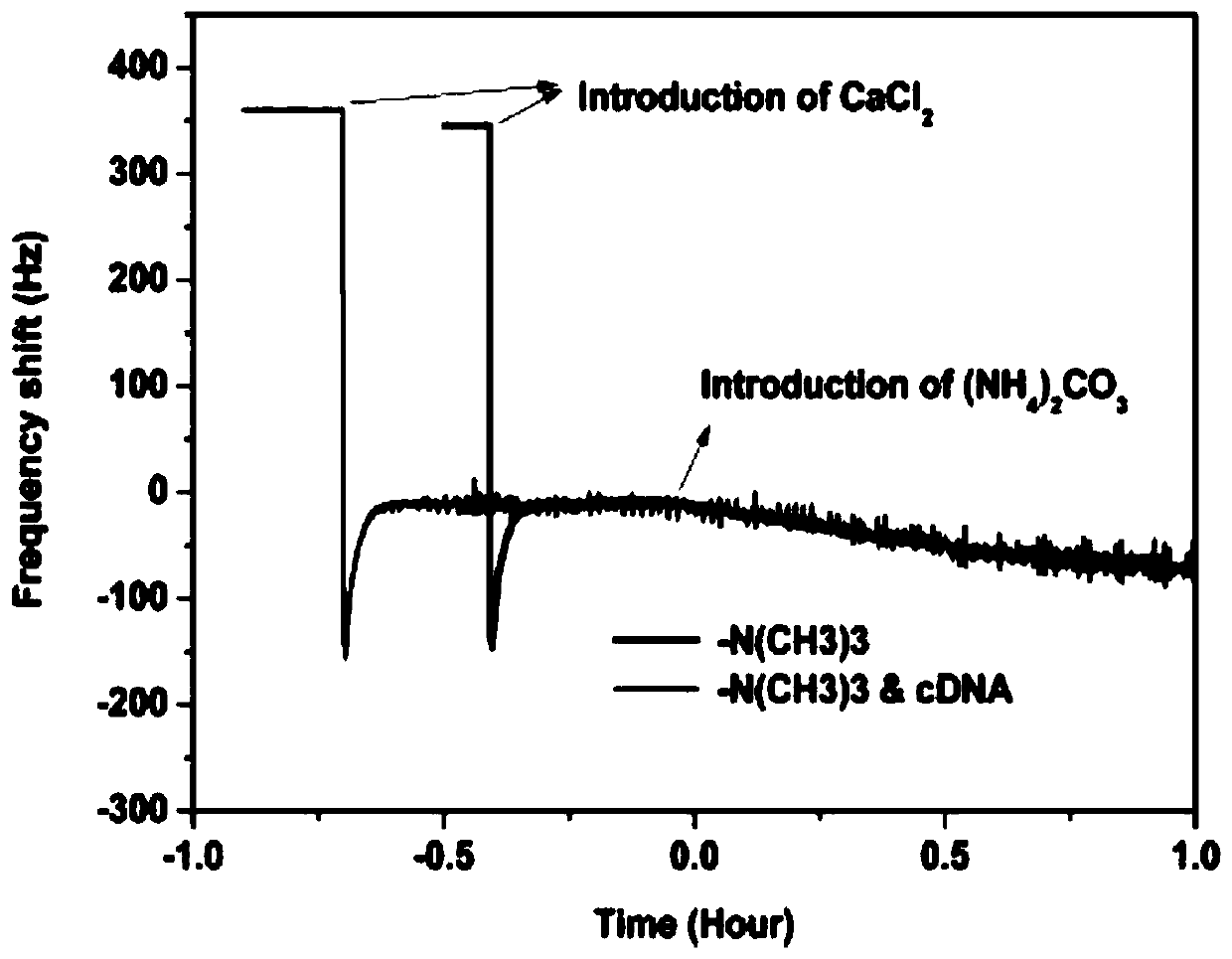
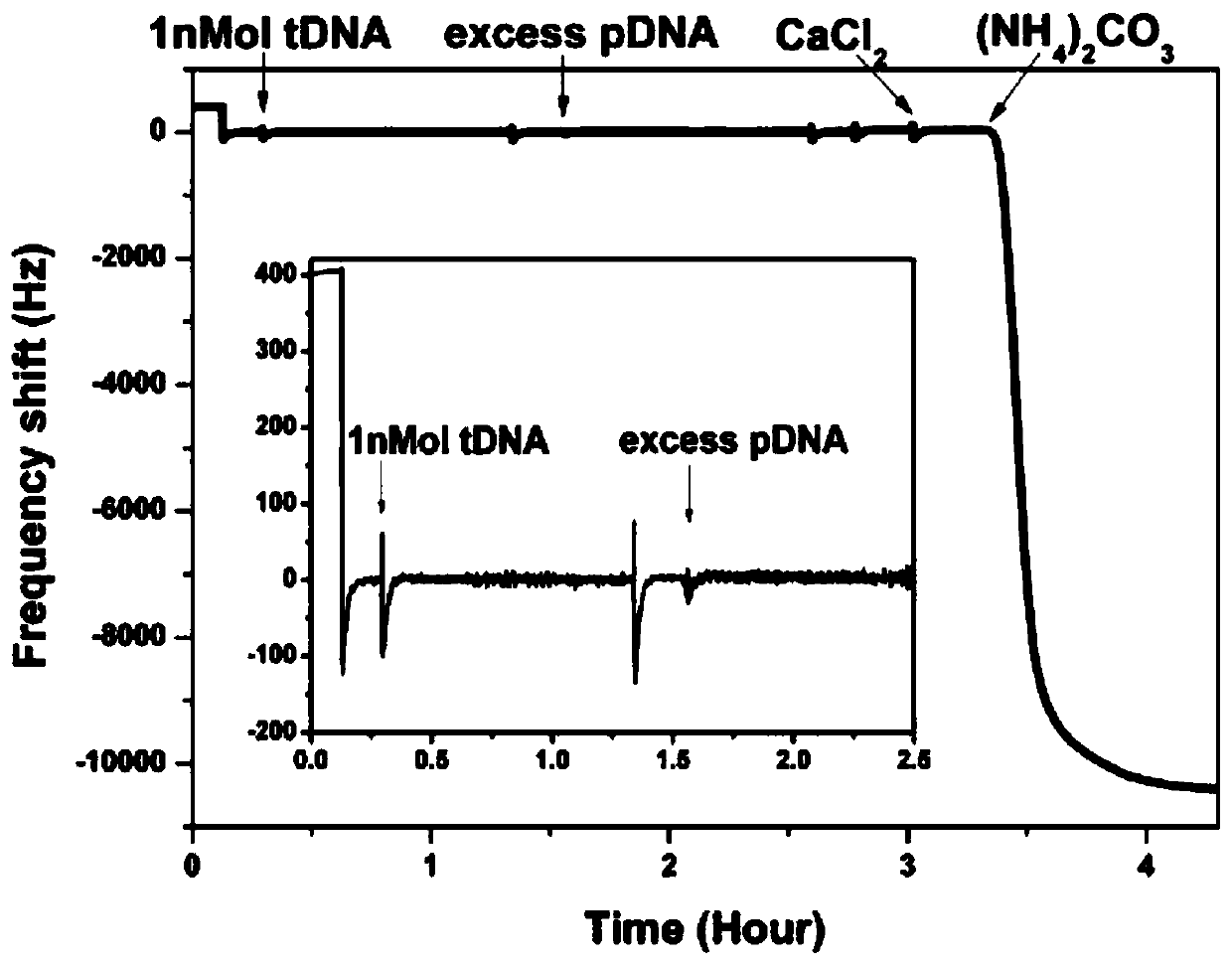
[0066] The capture of tDNA, the hybridization of pDNA and the in-situ growth of calcium carbonate crystals can all be monitored in real time by the quartz crystal microbalance sensor. The frequency of the whole process changes as figure 2 Shown. It can be seen from the inset that because the quality of DNA is very small, there is no obvious frequency change when the cDNA concentration is 1 nM, and there is no obvious frequency change after pDNA hybridization. But when ammonium carbonate is added, the crystallization process begins to occur, and a very obvious frequency change can be observed at this time. The frequency change exceeds 10000 Hz, which is about 1000 times the signal change when directly detecting DNA. This obvious frequency change indicates that crystallization is a good way to amplify the signal of a quartz crystal microbalance.
Embodiment 3
[0067] Example 3 Detection of DNA by selective crystallization method of quartz crystal Weitian plain
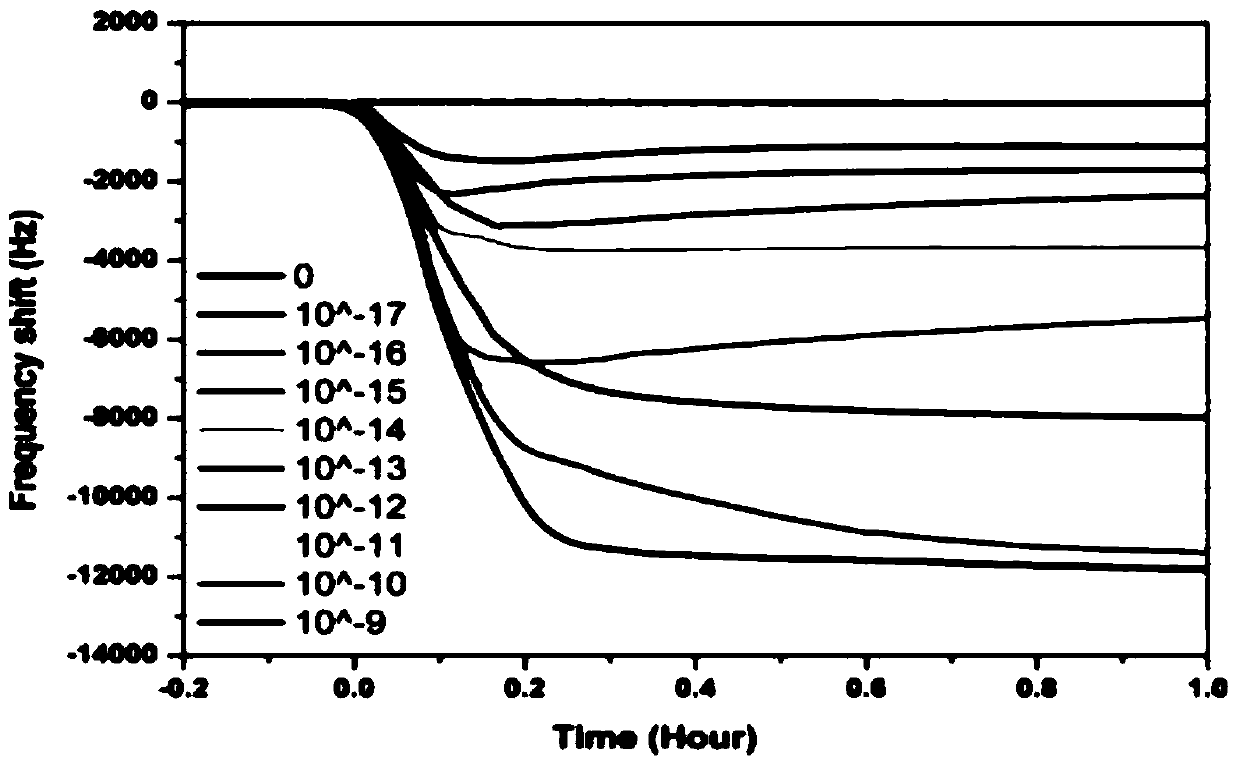
[0068] The in-situ selective crystalline quartz crystal microbalance response system is to detect and record a series of different concentrations of tDNA in real time. The results are as follows image 3 Shown. It was found that the higher the tDNA concentration, the greater the frequency change. The experiment of each concentration was repeated three times. The linear working curve between the quartz crystal microbalance signal response and the logarithm of the DNA concentration is as follows Figure 4 Shown. The linear regression satisfies the equation y=-21044.13-117073㏒c, and its correlation coefficient R 2 Is 0.997, where y is the frequency change of the quartz crystal microbalance sensor during the surface crystallization process, and c is the concentration of DNA. The linear range of DNA detection concentration is very wide, from 10aM to 1nM, and its minimum detection li...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com