Primer set for simultaneously detecting three kinds of microsporidia and application of primer set
A technology of microsporidia and primer sets, which is applied in recombinant DNA technology, microbial determination/inspection, resistance to vector-borne diseases, etc., can solve the problems of complex microsporidia process, low sensitivity and low specificity, etc. Simplified steps, broad application prospects, good sensitivity and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1, Establishment of Triple PCR of Enterocystis prawn hepatica, Enterocystis artemia and Microsporidia river crab 1. Materials and methods
[0031] 1. Samples: The positive samples of Enterocystis prawns, Enterocystis artemia and Microsporidium river crabs were all collected from Yingkou and Panjin, Liaoning, and separated by the laboratory.
[0032] 2. Reagents: DNA extraction kit was purchased from Tiangen (Beijing) Biochemical Technology Co., Ltd.; PCR Mix and PCR product recovery kit were purchased from Nanjing Novizan Biotechnology Co., Ltd.
[0033] 3. DNA extraction of three microsporidia
[0034] (1) Cut no more than 30 mg of tissue materials containing three types of microspores, put them into centrifuge tubes containing 200 μl GA buffer solution, and vortex for 15 seconds.
[0035] (2) Add 20 μL of Proteinase K (20mg / ml) solution, vortex to mix, and briefly centrifuge to remove water droplets on the inner wall of the tube cap. Place at 56°C until the ...
Embodiment 2
[0062] Embodiment 2, the specificity experiment of above-mentioned primer set detection sample
[0063] 1. Single primer specificity experiment
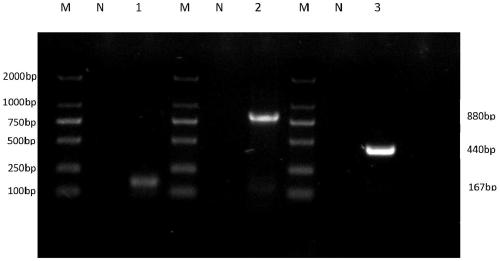
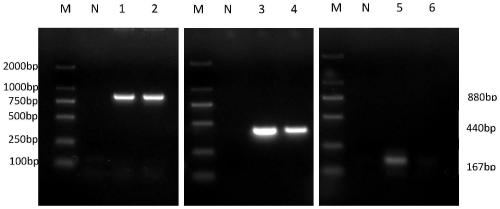
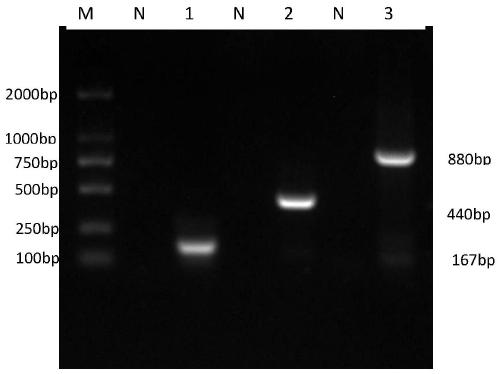
[0064] The three microsporidian DNA templates were mixed in equal proportions, and a pair of primers were added for PCR amplification to test the specificity of the primers. The results showed that only the gene corresponding to the specific primers could be amplified. After mixing the DNA of the three microsporidia, PCR amplification was performed with a single primer, and the results were as follows: image 3 As shown, each pair of primers can amplify the corresponding target gene fragment, but cannot amplify other gene fragments.
[0065] 2, 3 pairs of primer specificity experiments
[0066] After mixing the DNA of the three microsporidia, three pairs of specific primers from the above primer group were added to the sample at the same time, and PCR amplification was performed at the same time. The results showed that the gene fr...
Embodiment 4
[0067] Embodiment 4, the sensitivity experiment of above-mentioned primer set detection sample
[0068] The concentration of the PCR products recovered by gel cutting in Example 1 was measured, and the copy number of each sample was calculated according to the DNA copy number calculation formula. The results are shown in Table 2.
[0069] Table 2
[0070]
[0071] Copy number = (6.02×10 14 ×concentration (ng / μL) / (DNA length×660)
[0072] The copy numbers of the three microsporidian DNA templates were diluted to approximately 5.7×10 8 , 5.7×10 7 , 5.7×10 6 , 5.7×10 5 , 5.7×10 4 , 5.7×10 3 , 5.7×10 2 , 5.7×10 1 , the three DNA templates with the same copy number are mixed in equal proportions. Take 2 μL from the mixture as a template for PCR amplification.
[0073] The mixed sample was proportionally diluted according to the above method and used in the multiple PCR method established above to test the sensitivity of the established method. The results are as follo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com