Application of dideoxynucleoside phosphoramidite monomer containing succinimide structure in recognition of mismatched bases
A deoxynucleoside phosphoramidite and succinamide technology, applied in the field of non-natural oligonucleotide applications, can solve the problem of high cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
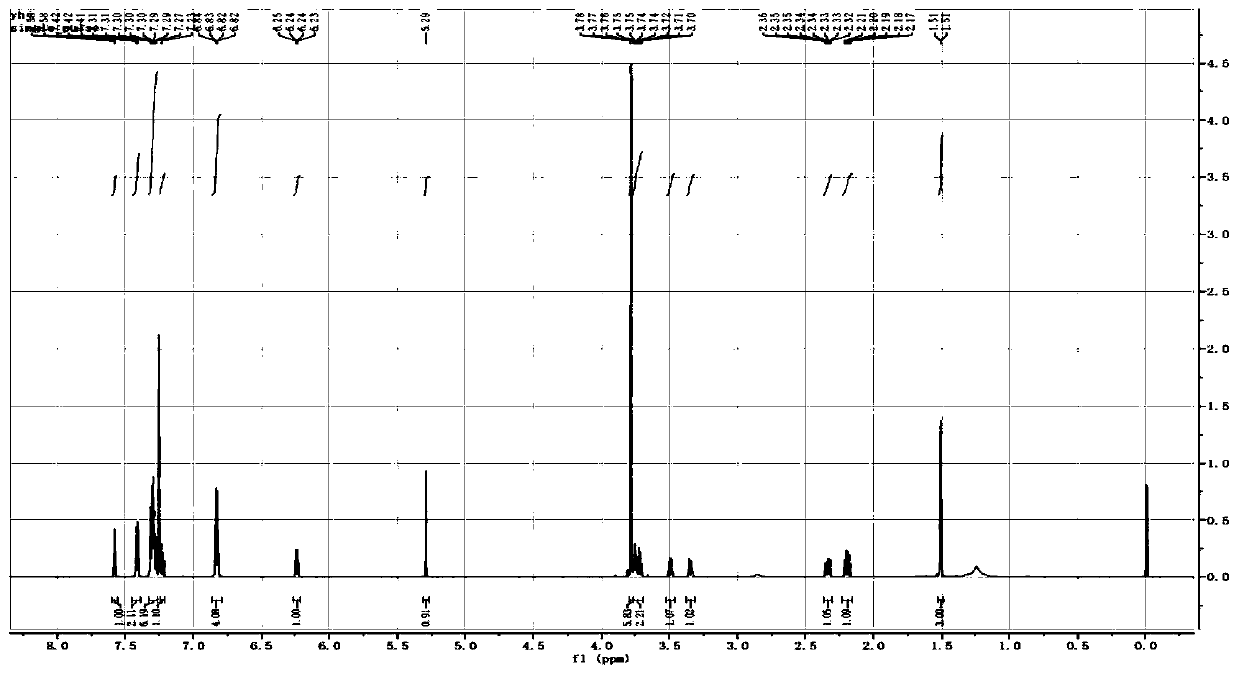
[0048] The present invention obtains a modification reagent of a non-natural oligonucleotide, that is, a deoxythymidine-deoxythymidine phosphoramidite monomer containing a succinamide structure, the chemical structure of which is as follows:
[0049]
[0050] The synthetic route of this monomer is as follows:
[0051]
[0052] Specific steps:
[0053] (a) Dissolve 10mmol zivudodine in 50mL anhydrous pyridine, add 10-20mmol 4,4-dimethoxytriphenylchloromethane in 10mL pyridine solution at room temperature, stir at room temperature for 1-12h; the reaction solution is concentrated After removing the solvent, add 50mL tetrahydrofuran, 1mL water and 1mL triethylamine, react at 85°C for 1-8h under the protection of nitrogen, concentrate to remove the organic solvent; Wash with water and dry over anhydrous sodium sulfate; the crude product can be directly used in the next step;
[0054] Dissolve 10mmol of the above-prepared product 5-(4,4-methoxytriphenylmethyl ether)-3-amino-...
Embodiment 2
[0070] Use of deoxythymidine-deoxythymidine phosphoramidite monomers containing succinamide structure for DNA synthesis
[0071] The synthetic route is as follows:
[0072]
[0073] Specific steps:
[0074] (d) 800 mg of double thymidine reagent (deoxythymidine-deoxythymidine phosphonamidite monomer containing succinamide structure) was dissolved in 3 mL of anhydrous acetonitrile, and transferred to the end group modification position of ABI394 nucleic acid synthesizer under nitrogen protection in the reagent bottle. Input nucleic acid sequence: 5'-TTC CAC TTA CCA GAT TGA TT-3', perform synthesis on the order of 1 μmol;
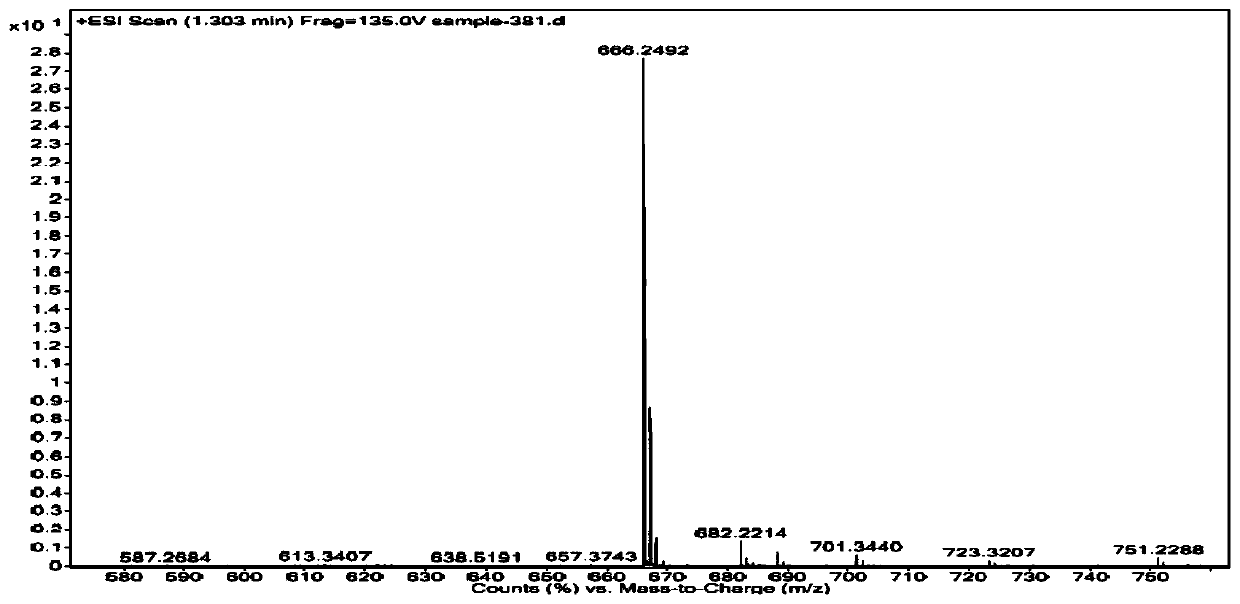
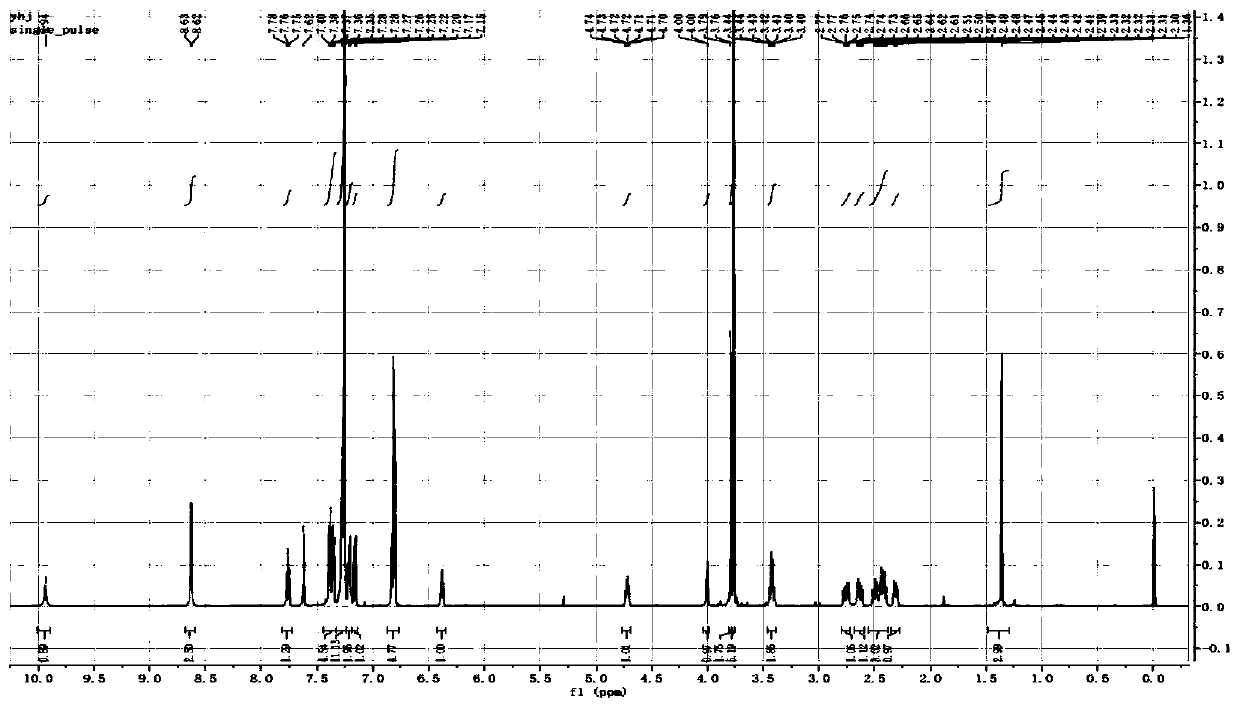
[0075] (e) After the synthesis is completed, the solid phase carrier is taken out, dispersed in 1mL 5-32% methylammonia aqueous solution, heated at 55-75°C for 1-4h, centrifuged to remove insoluble matter, and the solution is concentrated to obtain the crude product; the target product is obtained by Separation by HPLC and characterization of structural...
Embodiment 3
[0080] Application of Double Thymidine Reagent Containing Succinamide Structure in Construction of Resistant Enzyme Cutting Artificial Nucleic Acid
[0081] 1. A synthetic method for modifying the 5' end of an artificial nucleic acid with a double thymidine reagent containing a succinamide structure
[0082]
[0083] 2. A synthetic method for modifying the 3' end of an artificial nucleic acid with a double thymidine reagent containing a succinamide structure
[0084]
[0085] A method for synthesizing an enzyme-resistant artificial nucleic acid, the specific steps of which are:
[0086] Step a: Dissolve 800 mg of the reagent in 3 ml of anhydrous acetonitrile, and transfer it to the terminal modification site reagent bottle of the ABI 394 nucleic acid synthesizer under nitrogen protection. Input nucleic acid sequence: 5'-TTC CAC TTA CCA GAT TGA TT-3', perform synthesis on the order of 1 μmol.
[0087] Step b: After the synthesis is completed, the solid phase carrier is ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com