Method for quickly building CRISPR gene editing liver cancer cell strain and cell strain
A liver cancer cell, gene editing technology, applied in the field of gene editing targeting tumor cells, can solve problems such as unsatisfactory efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
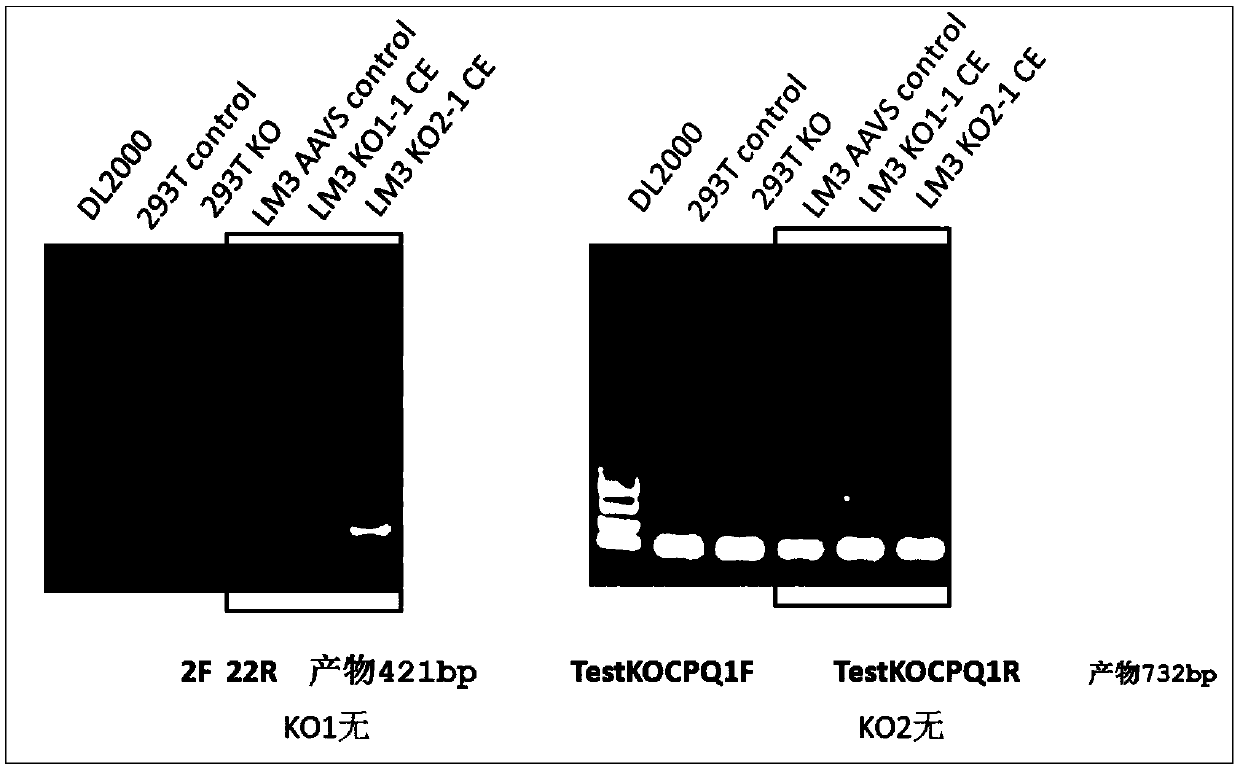
[0053] Example 1. Using optimized CRISPR / Cas9 technology to construct LM3 liver cancer cell line with knockout of CPQ gene
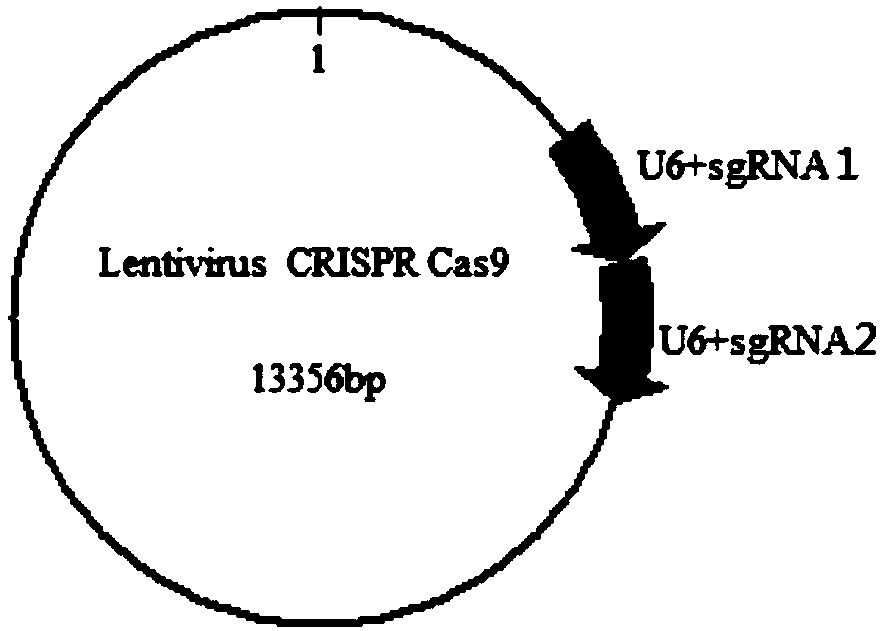
[0054] (1) With the Lentivirus V2 vector (Zhang Feng Lab, MIT) as the basic skeleton, the genome sequence of the human CPQ gene was downloaded from NCBI and the structure was analyzed to find the transcription start region, and a pair was selected based on the online design of this region sgRNA, its upstream and downstream sgRNA sequences are as follows:
[0055] sgRNA1 CTTATCTTCGCATTTTTCGG(TGG)
[0056] sgRNA2 CAGAAGTGCCAATCGCTCAT(AGG)
[0057] Among them, the three bases in brackets are the PAM domain (NGG)
[0058] The sequences of four primers were designed as follows:
[0059] KOCPQ primer I (sgRNA1 sequence is underlined)
[0060] 5′-CGTCTCGCACCGCTTATCTTCGCATTTTTCGGgttttagagctaGAAAtag-3′
[0061] KOCPQ primer II
[0062] 5′-CAAAAAAGCACCGACTCGGTGCCACTTTTTTC-3′
[0063] The first PCR uses primers primer I and primer II to obtain a PCR product ...
Embodiment 2
[0097] Embodiment 2, the establishment of stably knocking out the MTDH gene LM3 cell line, the specific implementation steps are as follows:
[0098] (1) Construction of a plasmid for knocking out the MTDH gene. The two pairs of sgRNA sequences are:
[0099] first pair:
[0100] MTDH primer I 5'-CGTCTCGCACCGGAAATGCTCTCGGTCGGCCTGTTTTAGAGCTAGAAATAG-3';
[0101] MTDH primer IV
[0102] 5'-CGTCTCCAAACAGCGCCCGCAAAAAGCGGAGGCGGTGTTTCGTCCTTTCCAC-3'.
[0103] Second pair:
[0104] MTDH 2primer I
[0105] 5'-CGTCTCGCACCGAAATGGGCGGACTGTTGAAGGTTTTTAGAGCTAGAAATAG-3';
[0106] MTDH 2primer IV
[0107] 5'-CGTCTCCAAACATAGTGGATGGGTGGTAAAAGCGGTGTTTCGTCCTTTCCAC-3'.
[0108] After extracting the knockout MTDH gene recombinant plasmid obtained in the present invention and measuring the concentration of the plasmid with NanoDrop1000, the integrity of the plasmid is detected by electrophoresis.
[0109] (2) 24 hours before cell transfection, inoculate a cell density of 8×10 in a 100 mm cultur...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com