Method for producing 2'-fucosyllactose by using escherichia coli
A technology of fucosyllactose and Escherichia coli, which is applied in the field of microorganisms and can solve the problems of low efficiency of 2'-fucosyllactose
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] 1. Knockout method
[0052] Through the NCBI genome database, the operon sequence of Escherichia coli colanic acid was obtained, and PCR primers were designed. Using the bacterial liquid of Escherichia coli S17-3 as a template, the upstream and downstream of the wcaJ gene shown in SEQ ID NO. Each sequence was amplified by 300bp to obtain adjacent homology arm DNA of wcaJ. The primers for amplifying upstream homologous fragments are:
[0053] Ant-F:AGCGGTGAGATCAACAGCAAACTGG (SEQ ID NO.8), and
[0054] Ant-R:TAGGAACTTCGAAGCAGCTCCAGCCTACACAATCGCTCCGTTGTTCCTGTTATTAGCCCCTTACCCG (SEQ ID NO.9), the PCR reaction system for amplifying the upstream homology arm fragment by primer pair Ant-F, Ant-R is as follows:
[0055]
[0056] The PCR program is as follows:
[0057] 1) 95°C, 3min;
[0058] 2) 95°C, 15sec;
[0059] 3) 56°C, 15sec;
[0060] 4) 72°C, 2.5min;
[0061] 5) Cycle from 2) to 4), 30 times;
[0062] 6) 72°C, 5min;
[0063] 7) Store at 4°C.
[0064] Similarl...
Embodiment 2
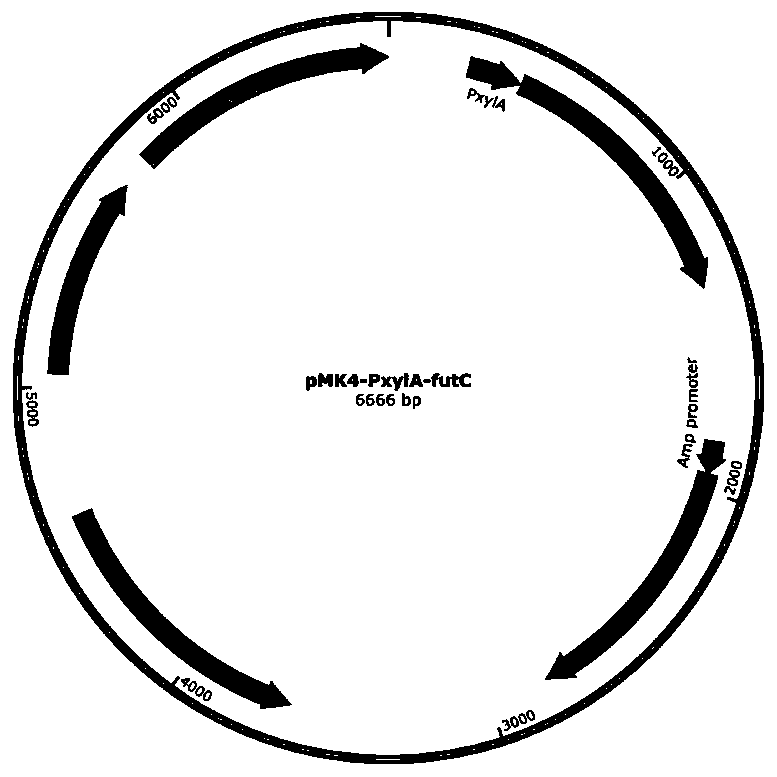
[0086] Embodiment 2: Construction of recombinant plasmid pMK4-pxylA-FutC
[0087] First, the expression element of the vector was obtained from the plasmid pMK4-pxylA-GFP stored in the laboratory by PCR amplification, and the amplification primers were:
[0088] pMK4-F:ATGAATTCACTGGCCGTCGTTTTACAAC (SEQ ID NO. 3)
[0089] pMK4-R: TTTTTATTCCTCCTTGTTCCCGGGTTGATT (SEQ ID NO. 4)
[0090] The PCR reaction system of the pMK4 expression element of the amplified fragment pMK4 by the primer pair pMK4-F, pMK4-R is as follows:
[0091]
[0092] The PCR program is as follows:
[0093] 1) 95°C, 3min;
[0094] 2) 95°C, 15sec;
[0095] 3) 56°C, 15sec;
[0096] 4) 72°C, 2.5min;
[0097] 5) Cycle from 2) to 4), 30 times;
[0098] 6) 72°C, 5min;
[0099] 7) Store at 4°C.
[0100]Nucleotide sequences related to the expression of fucosyltransferase FutC were generated by Jinweizhi Biotechnology Company using DNA chemical synthesis and constructed into the cloning vector pUC19.
[0101...
Embodiment 3
[0105] Example 3: Fermentative production of 2'-fucosyllactose by recombinant Escherichia coli under initial low pH conditions
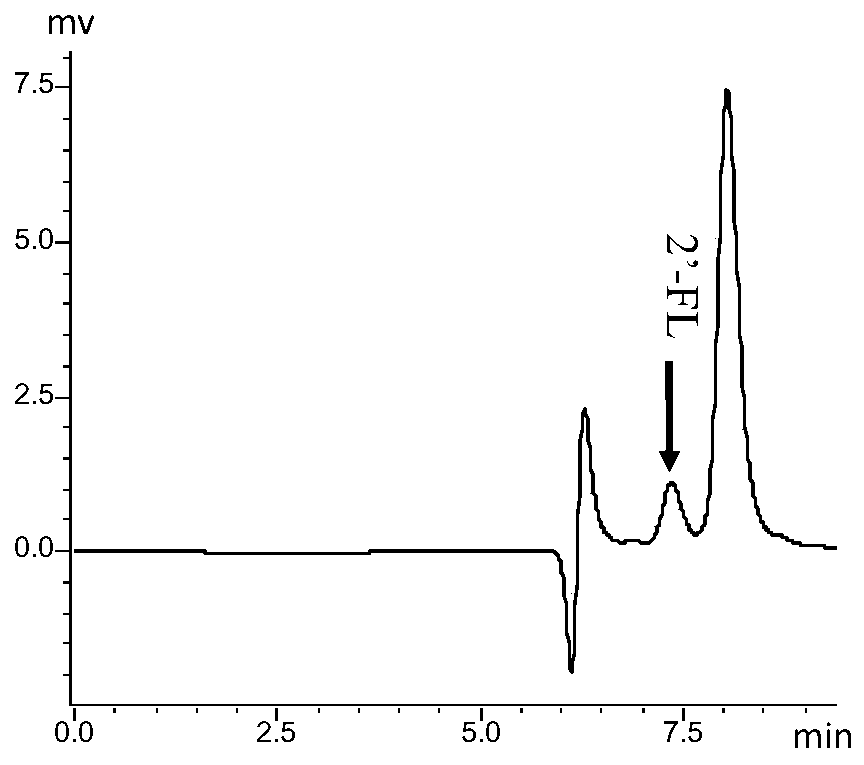
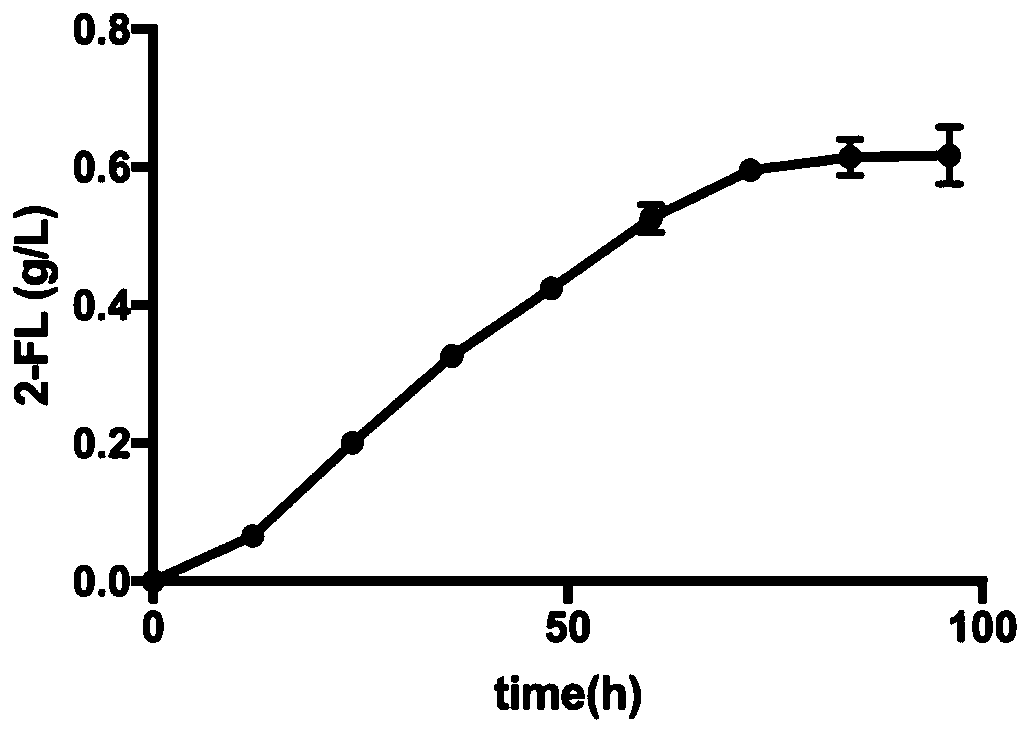
[0106] Use the recombinant Escherichia coli mentioned above as the production strain for shake flask fermentation: Pick the recombinant Escherichia coli with an inoculation loop and inoculate it into a test tube containing 3 ml of LB, culture overnight at 37°C on a shaker at 200r / min. Inoculate the Escherichia coli overnight cultured with 1% inoculum into a 500mL shake flask containing 50mL liquid LB medium, add 10g / L lactose, 20g / L glycerol and 50ul of 100ug / ml ampicillin Antibiotics were also fermented at 37°C and 200r / min in a shaker. The initial pH of the medium was controlled at pH 4.5 using 3M hydrochloric acid. The shake flask was fermented continuously for 96 hours, and the 2'-FL production was measured by taking samples at intervals of 12 hours.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com