Method for modifying ubiquitin and inhibiting ubiquitination pathway
A technology of ubiquitination and ubiquitin, which is applied in the field of life science and microbial protein function and application, and can solve the problem that the modification is not the final product
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Expression and purification of CteC recombinant protein
[0027] 1. Construction of expression plasmid
[0028] Using the genomic DNA of Chromobacterium violaceum ATCC 12472 strain as a template, primers F1: GATAGGATCCATGCTATTTTTCACCGGTCTGC (SEQ ID NO. 1) and R1: GATACTCGAGTTAGACCGACGCCAACTCCTG (SEQ ID NO. 2) were designed according to the gene sequence (CV_1647) on NCBI to obtain CteC by PCR amplification Full length target fragment.
[0029] The CteC full-length target fragment was constructed into the kanamycin-resistant prokaryotic expression plasmid pRSF-Duet-His-SUMO vector (the vector is in pRSF-Duet1) by restriction digestion (the restriction sites are BamH1 and Xho1) and ligation. (Novagen), pRSF-Duet1-His-SUMO-CteC is obtained by inserting SUMO protein after His tag. His tag protein can be affinity purified with Ni magnetic beads, and SUMO protein can be specifically recognized by Ulp1 enzyme for subsequent excision of tag protein.
[0030] 2. Protein expression
[0...
Embodiment 2
[0034] Example 2: Effector protein CteC on ADP ribosylation modification of ubiquitin molecules
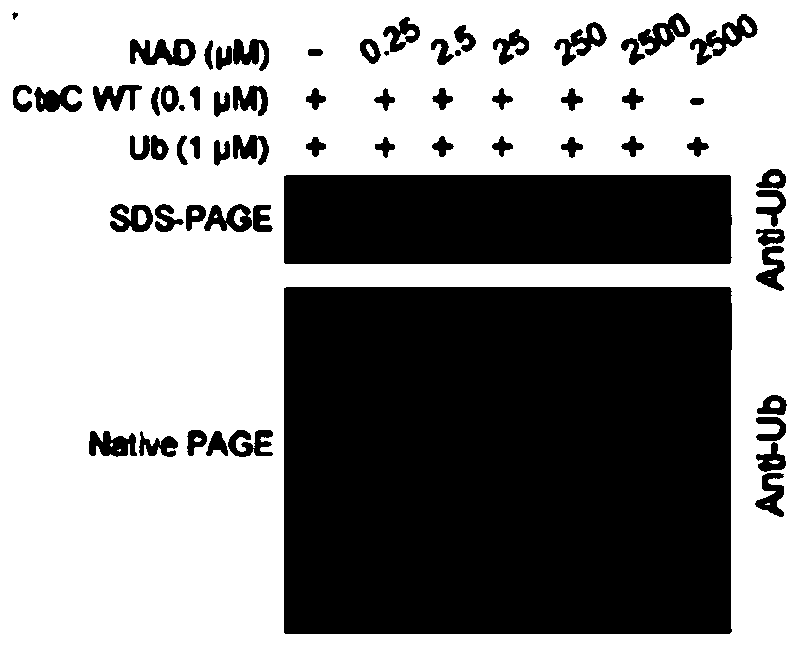
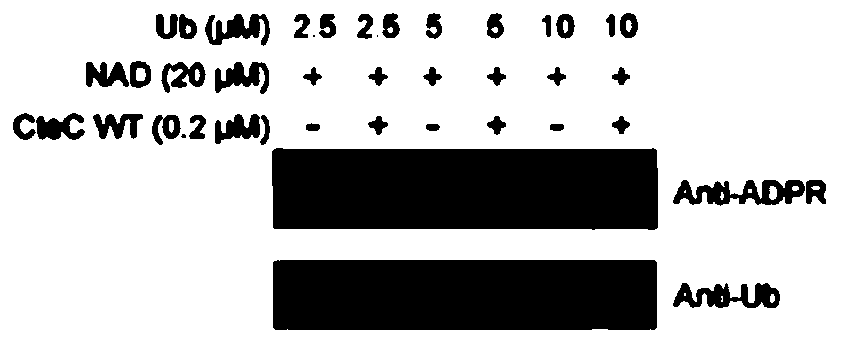
[0035] In a 15μL reaction system (the reaction buffer is 20mM HEPES pH 7.4, 150mM NaCl), add 1μM Ub (the protein purification method is the same as the reference Wan et al., 2019, doi:10.1038 / s41564-019-0454-1) And 0.1 μM of the CteC protein purified in Example 1, set the NAD concentration gradients to 0, 0.25, 2.5, 25, 250, 2500 μM, and after reacting at 37°C for 30 minutes, run 15% SDS-PAGE and 8% Native PAGE, respectively If Ub undergoes a modification reaction, it can migrate upward on 15% SDS-PAGE gel, and migrate downward on 8% Native PAGE. In vitro biochemical experiments show that the modification of Ub by CteC needs to rely on NAD as a ligand ( figure 2 ), and the modified Ub can be recognized by the ADPr antibody (MABE1016, Millipore) ( image 3 ). Through sequence alignment analysis, it is found that CteC is similar to some ARTs and has a conservative "RSE motif" ( Figu...
Embodiment 3
[0038] Example 3 The effect of the effector protein CteC on the ADP ribosylation modification of ubiquitin molecules on the ubiquitination pathway and host cells.
[0039] Ubiquitin molecules participate in various signal pathways after forming different forms of ubiquitin chains through cascade reactions. Using different E2 and E3 will form a specific form of ubiquitin chain. In this example, UbcH5c and IpaH3 were used to synthesize K48 chain, and Uev1a, Ubc13 and TRAF6 were used to synthesize K63 chain. (Refer to Wanet al., 2019, doi: 10.1038 / s41564-019-0454-1 for protein particle information and purification methods involved in this embodiment) The experimental results show that the efficiency of CteC-modified Ub in the formation of K48 and K63 chains in vitro is greatly reduce( Picture 9 ).
[0040] The NF-κB signaling pathway plays an important role in the cellular immune response, and the degradation of IκBα is an indicator to detect whether this pathway is activated. The...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com