Method for predicting affinity of HLA type I molecule and polypeptide
A prediction method, affinity technology, applied in the field of bioinformatics
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0071] The present invention will be specifically introduced below in conjunction with the accompanying drawings and specific embodiments.
[0072] An optimization process of an affinity prediction method for HLA class I molecules and polypeptides, comprising:
[0073] 1. Dataset selection:
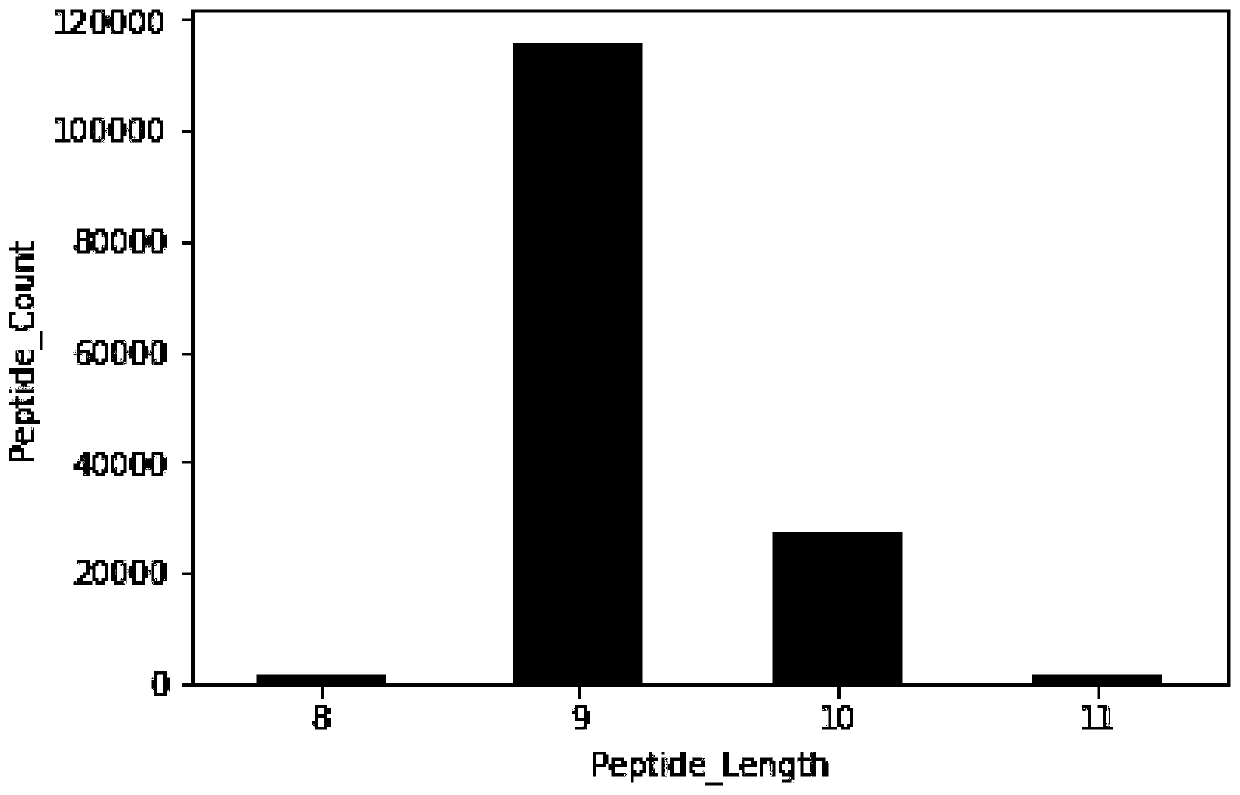
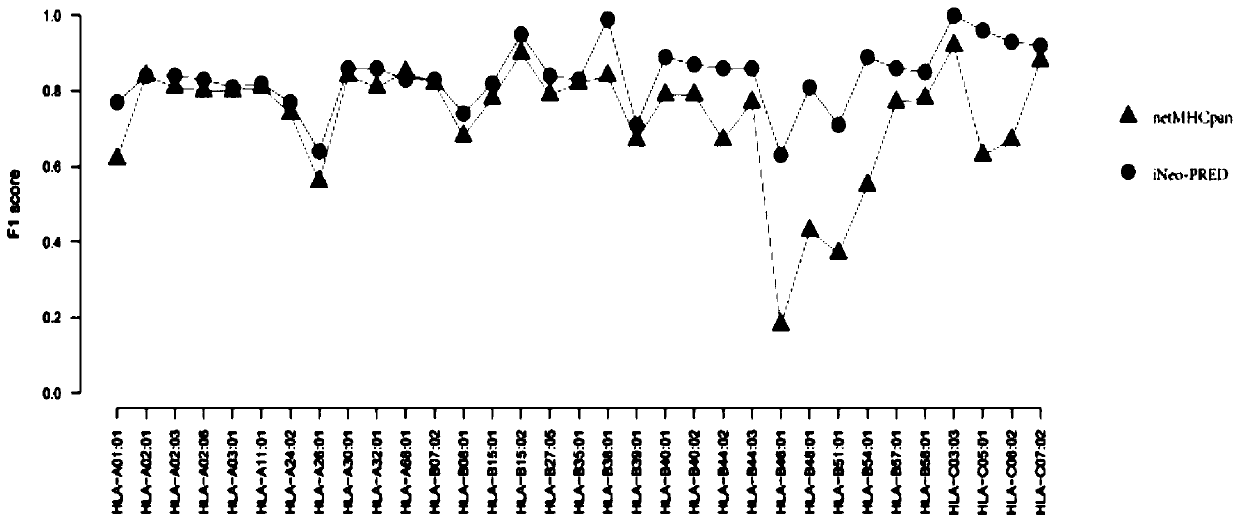
[0074] Due to the large difference in the amount of training data corresponding to different typing, and the similarity between the peptides corresponding to different typing, the prediction results of the previous software are biased, and the effect is the same for typing with a small amount of data. very bad. Through a large number of pre-tests, it is found that if you want to use machine learning methods to build models and get better learning results, each typing needs at least 1000 polypeptide sequences as training data, so the present invention uses typing with a data volume greater than 1000 for For the construction of the machine learning model, for typing with a small amount of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com