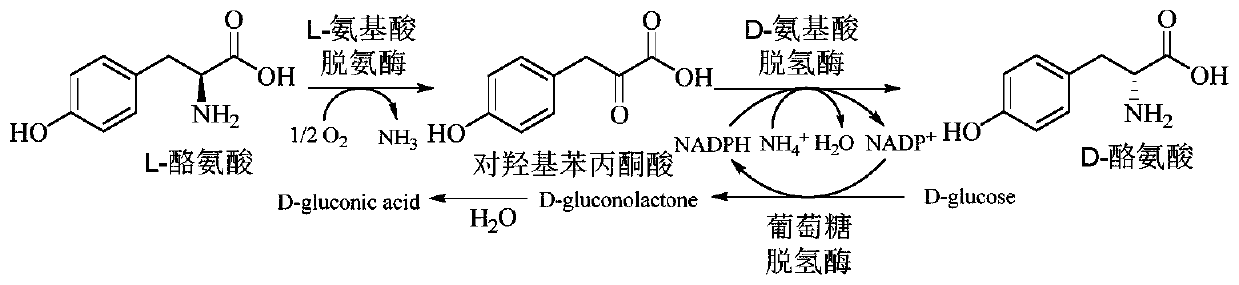
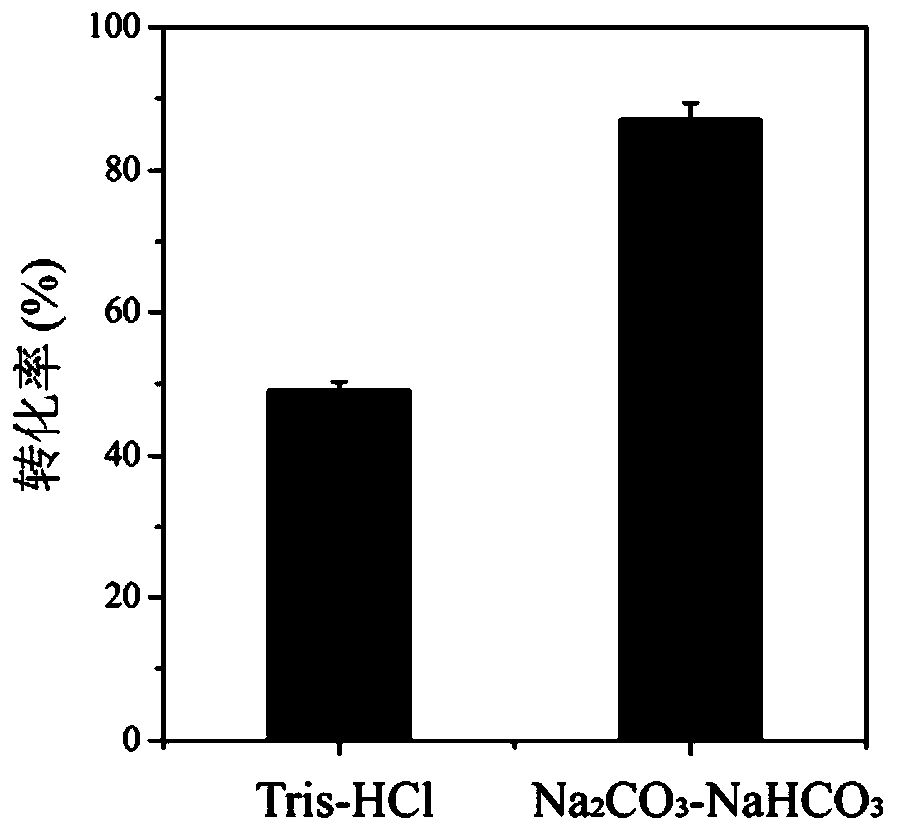
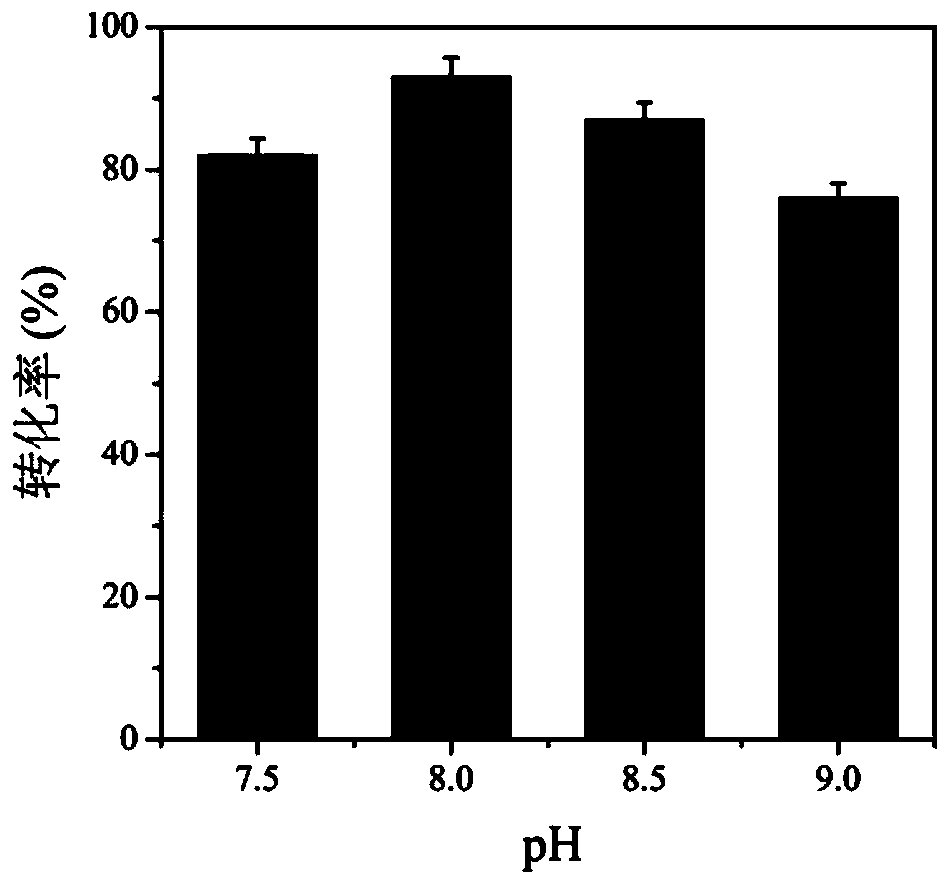
Method for improving production efficiency of D-tyrosine
A tyrosine and amino acid technology is applied in the field of improving the production efficiency of D-tyrosine, which can solve the problems of low conversion rate of L-tyrosine, and achieve the effects of high optical purity, strong specificity and low cost of products.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Embodiment 1: Construction of recombinant plasmid pRSFDuet-1-PmLAAD
[0032] The artificially synthesized PmLAAD gene (nucleotide sequence shown in SEQ ID NO: 1) containing SacI and SalI restriction sites through codon optimization, the PmLAAD gene and the plasmid pRSFDuet- Digest at 137°C for 2 hours, use T 4 The PmLAAD gene and plasmid pRSFDuet-1 after enzyme digestion and gel recovery were ligated with ligase for 10 hours at 16°C, and the ligated products were transformed into E.coli JM109 competent cells by chemical transformation, and cultured on LB plates containing kanamycin for 12 hours , and verify the colonies grown on the plate by PCR. Positive transformants were selected and inoculated in LB medium, cultured at 37°C for 12 hours, and plasmids were extracted. After sequencing verification, the recombinant plasmid pRSFDuet-1-PmLAAD was constructed.
Embodiment 2
[0033] Example 2: Construction of recombinant plasmid pACYCDuet-1-CgDAPDH
[0034] The artificially synthesized CgDAPDH gene (nucleotide sequence shown in SEQ ID NO: 2) containing EcoRI and SalI restriction sites through codon optimization, the CgDAPDH gene and the plasmid pACYCDuet- 1 Digest for 2 hours at 37°C, use T 4Ligase ligated the CgDAPDH gene and plasmid pACYCDuet-1 after digestion and gel recovery at 16°C for 10 hours, and transformed the ligation product into E.coli JM109 competent cells by chemical transformation method, and cultured them on LB plates containing chloramphenicol for 12 hours. Colonies grown on the plate were verified by PCR. Positive transformants were selected and inoculated in LB medium, cultured at 37°C for 12 hours, and plasmids were extracted. After sequencing verification, the recombinant plasmid pACYCDuet-1-CgDAPDH was constructed.
Embodiment 3
[0035] Example 3: Construction of recombinant plasmid pACYCDuet-1-CgDAPDH-BmGDH
[0036] Using the genome of Bacillus megaterium as a template, with 5,ga agatct catgtataaagattagaagg3, as forward primer, with 5, ccg ctcgag ttatccgcgtcctgcttg3, the BmGDH gene (nucleotide sequence shown in SEQ ID NO: 3) is amplified for the reverse primer (the underlined part is the restriction site of BglII and XhoI respectively), and the restriction endonuclease BglII and XhoI are used Digest the BmGDH gene and the plasmid pACYCDuet-1-CgDAPDH constructed in Example 2 at 37°C for 2 hours, and use T 4 The BmGDH gene and the plasmid pACYCDuet-1-CgDAPDH after digestion and gel recovery were ligated with ligase at 16°C for 10 hours, and the ligated product was transformed into E.coli JM109 competent cells by chemical transformation, and cultured on LB plates containing chloramphenicol After 12 hours, the colonies grown on the plate were verified by PCR. Positive transformants were selected and ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com