Fragment combination, specific primer and application for identification of Rosaceae plant species
A species identification, Rosaceae technology, applied in the determination/inspection of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0126] Example 1, Screening Gene Loci and Primer Design for Identification of Rosaceae Plants
[0127] Collect a large number of measured Rosaceae chloroplast gene fragments and nuclear gene fragments to screen the initial data set of hypervariable gene fragments in Rosaceae plants, group the data sets according to gene fragments, and retain gene fragments containing at least 5 species, and use MAFFT software for comparison sequence, and then use BioEdit software to manually adjust the comparison results, and evaluate each gene fragment through the written python script. Finally, 7 gene fragments and 8 gene loci (matK, ITS-1, ITS-2, rbcL, trnH-psbA, atpB-rbcL, trnL, sbeI) were selected for the identification of Rosaceae plants.
[0128] Eight pairs of specific primers for the identification of Rosaceae plants were designed for the eight gene loci, and the specific information is shown in Table 1.
[0129] Table 1 Primer information
[0130]
Embodiment 2
[0131] Embodiment 2, establishment of detection method
[0132] 1. Extract the genomic DNA of the sample to be tested.
[0133] 2. Use the genomic DNA extracted in step 1 as a template, and use the primer pairs in Table 1 to perform PCR amplification to obtain PCR amplification products. The core codes matK and rbcL of the plant DNA standard barcode and the supplementary barcodes ITS and trnH- The primers corresponding to psbA are used for identification. If the primers corresponding to the above four barcodes cannot identify the species, other primers can be selected. For simple groups, a single or a combination of a small number of primer pairs can be selected for identification, and for complex groups, multiple primer pairs are required for joint identification. .
[0134] PCR amplification system (10μl): 10×PCR Buffer (Mg 2+ ) 1μl, 2mM dNTP 1μl, upstream primer 0.5μl, downstream primer 0.5μl, genomic DNA 1μl, 5U / μl Taq DNA polymerase 0.1μl, ddH 2 O 5.9 μl.
[0135] Eac...
Embodiment 3
[0141] Embodiment 3, actual detection
[0142] The DNA of all the samples in this example can be obtained from the Chinese Plant DNA Bank.
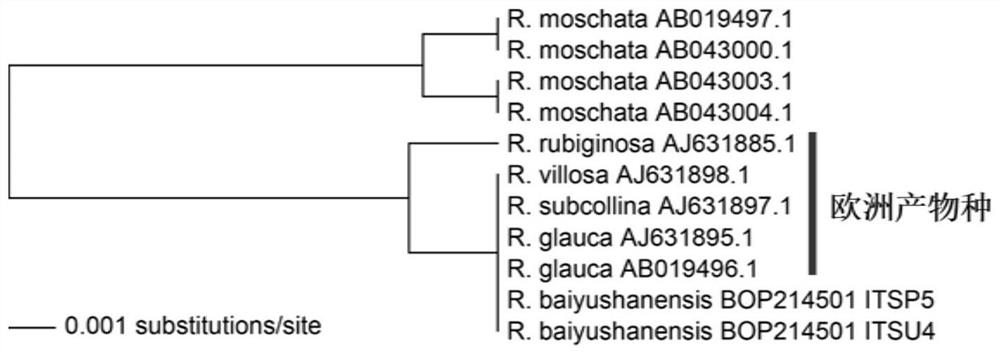
[0143] 1. Identify whether Rosa basilisk is a rare and endangered species
[0144] Samples to be tested: BOP214501 (Rose Rosaceae native to Lushun, used to identify whether it is a rare and endangered species).
[0145] The detection was carried out according to the method of Example 2, and the primer pair composed of Ros-ITS-1-F and Ros-ITS-1-R in Table 1 was used for PCR amplification. The sequencing result of the target fragment is shown in sequence 17 of the sequence listing.
[0146] The result is as figure 1 The topology is shown.
[0147] The results showed that Rosa basilicum escaped from European rose genetic resources.
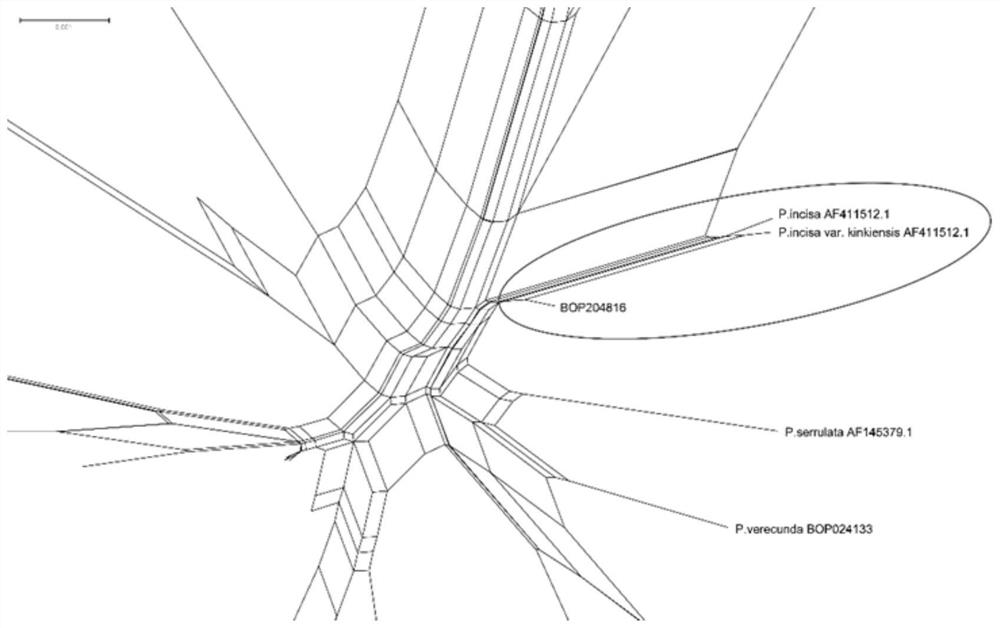
[0148] 2. Cherry Blossom Identification
[0149] Sample to be tested: a dry leaf of an unknown cherry blossom, numbered BOP204816.
[0150] The detection was carried out according to the method of Example...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com