Salmonella source tracing typing method based on gMLST (genome multilocus sequence typing) technology and application
A Salmonella, typing technology, applied in biochemical equipment and methods, microbial measurement/testing, and resistance to vector-borne diseases, etc., can solve the problem that no patent publications have been found, and the ability of MLST to distinguish cannot meet the needs of identifying non-outbreak pneumonia Cree To solve problems such as Burger bacteria strains, to achieve the effect of strong innovation and high resolution ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
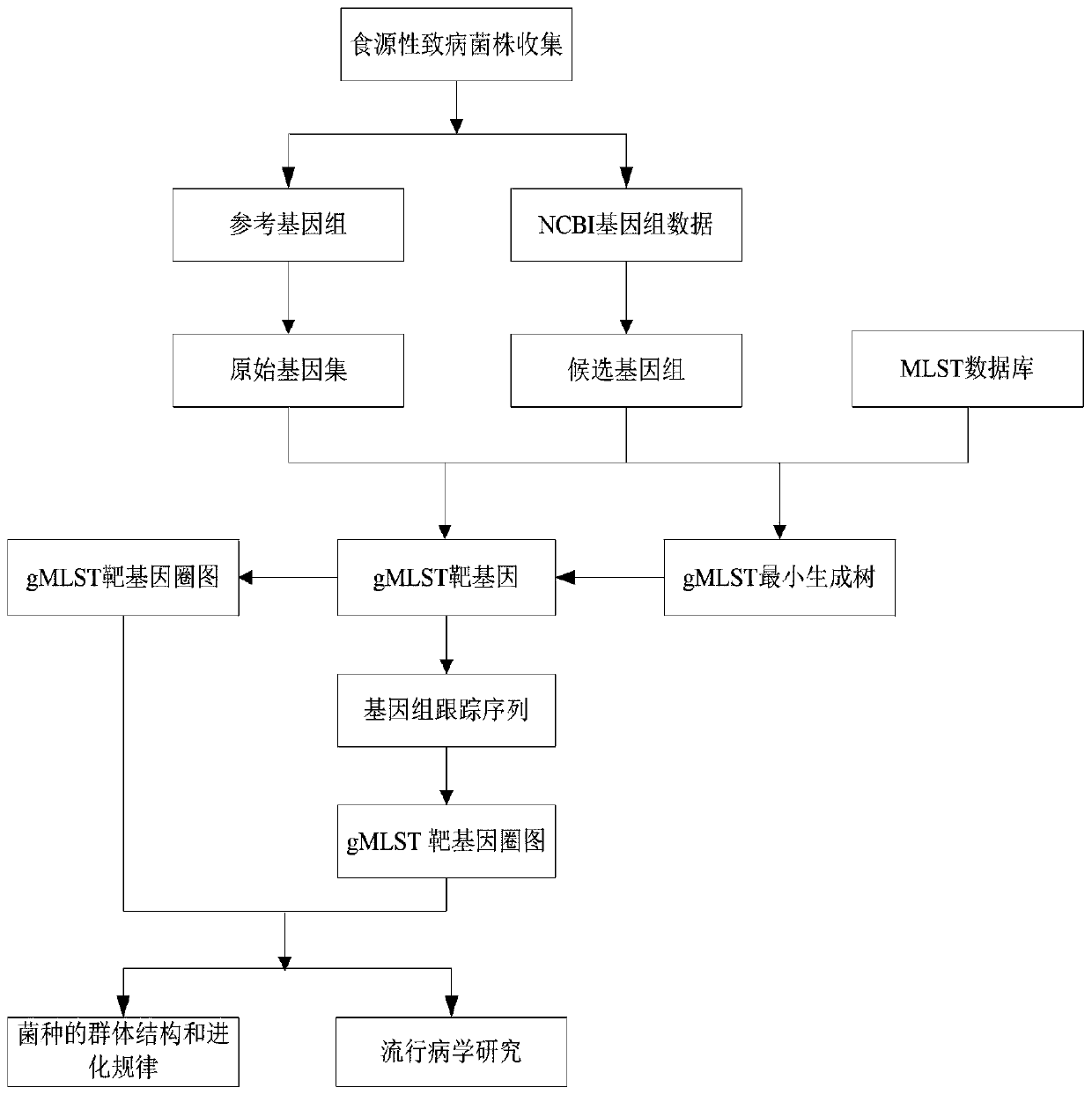
[0056] A method for traceability typing of Salmonella based on gMLST technology, the steps are as follows:
[0057] (1) Isolation and identification of Salmonella:
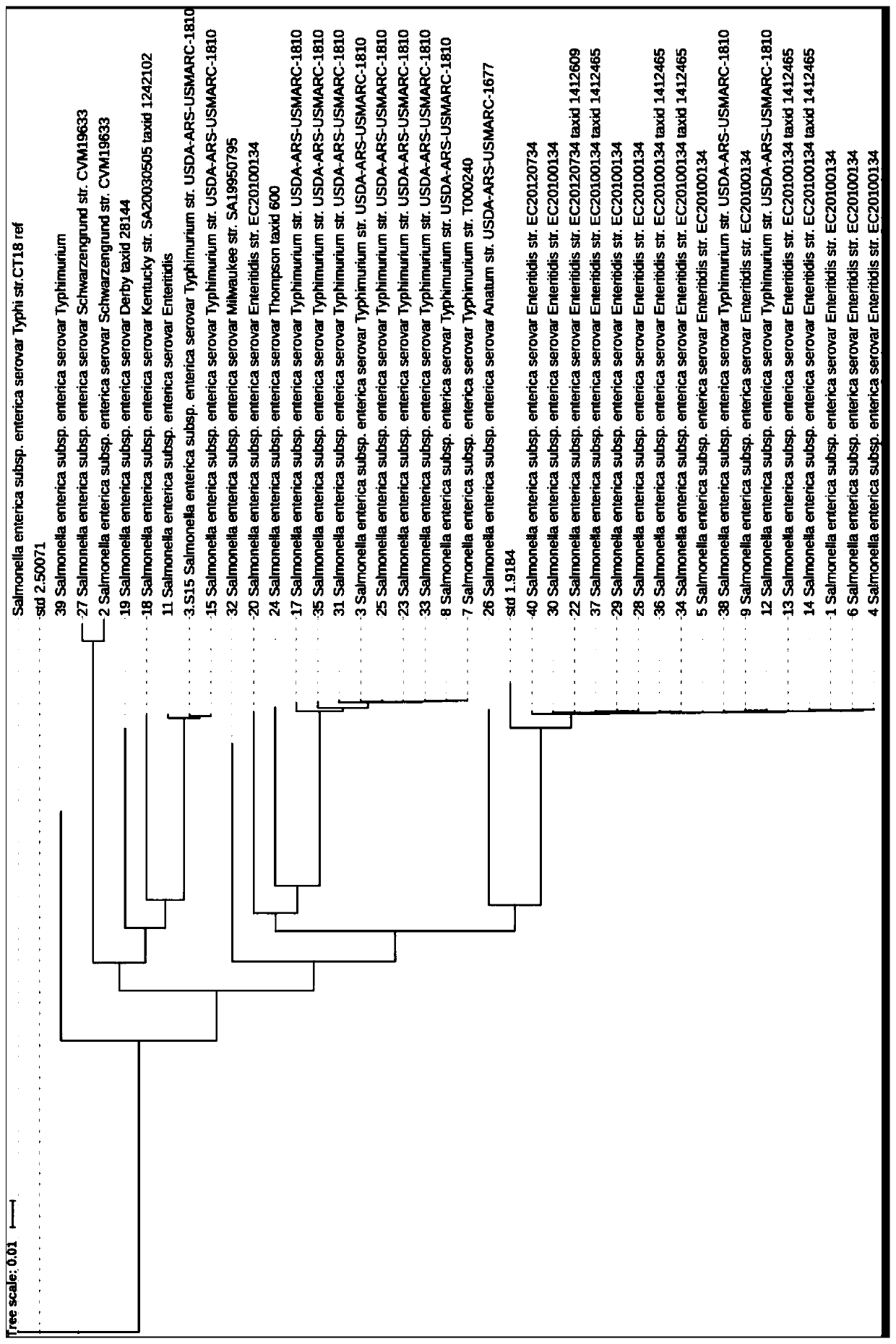
[0058] According to the national standard 4789.4-2016, more than 40 meat samples from the same area were collected for the isolation and identification of Salmonella. The 2 reference strains were the standard strains, which were Salmonella Enteritidis ATCC9184 and Salmonella Typhi CMCC 50071;
[0059] ⑵ DNA extraction, content detection:
[0060] Extract the DNA of the Salmonella sample isolated from the meat sample, melt the extracted DNA on ice, pipette, mix and centrifuge for detection; use agarose gel electrophoresis to detect the integrity of the sample and use it to construct a DNA library;
[0061] (3) Whole genome sequencing and g MLST typing
[0062] ① Whole-genome sequencing: de novo sequencing of the bacterial genome, constructing a library with qualified samples, using ultrasonic method to randomly i...
Embodiment 2
[0086] A method for traceability typing of Salmonella based on gMLST technology, the steps are as follows: (1) Isolation and identification of Salmonella.
[0087] According to the national standard 4789.4-2016, 285 meat samples collected in Beijing area were isolated and identified and the strains were preserved. 2 reference strains Standard strains are Salmonella enteritidis ATCC9184, Salmonella typhi CMCC 50071 (ATCC Culture Collection, USA)
[0088] (2) DNA extraction and content detection.
[0089] According to the extraction steps of the DNA extraction kit, the extracted DNA from 40 samples was melted on ice, then pipetted, mixed and centrifuged for detection. The integrity of the sample was detected by agarose gel electrophoresis (gel concentration 1%; voltage 150V, electrophoresis time 40min), and was used to construct a DNA library.
[0090] (3) Whole genome sequencing and g MLST typing
[0091] (1) Whole genome sequencing: bacterial genome de novo sequencing. The...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com