A method for predicting association relationship between metabolites and diseases based on a KATZ model
A technology of association relationship and prediction method, which is applied in the field of bioinformatics, can solve problems such as too large deviations, and achieve the effect of improving prediction performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
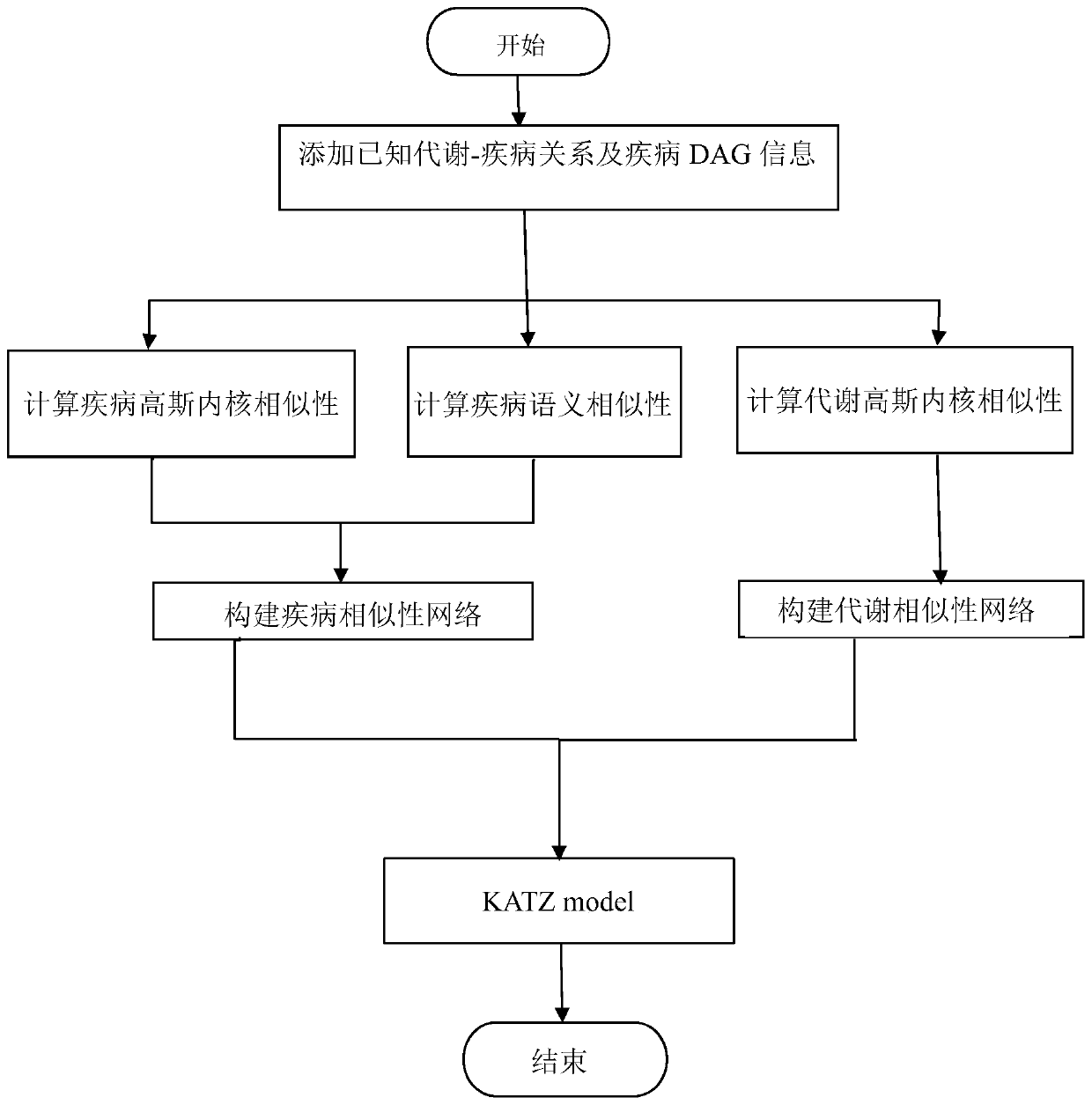
[0089] Taking the metabolite and disease network as an example, the steps of a method for predicting the relationship between metabolites and diseases based on the KATZ model are as follows:
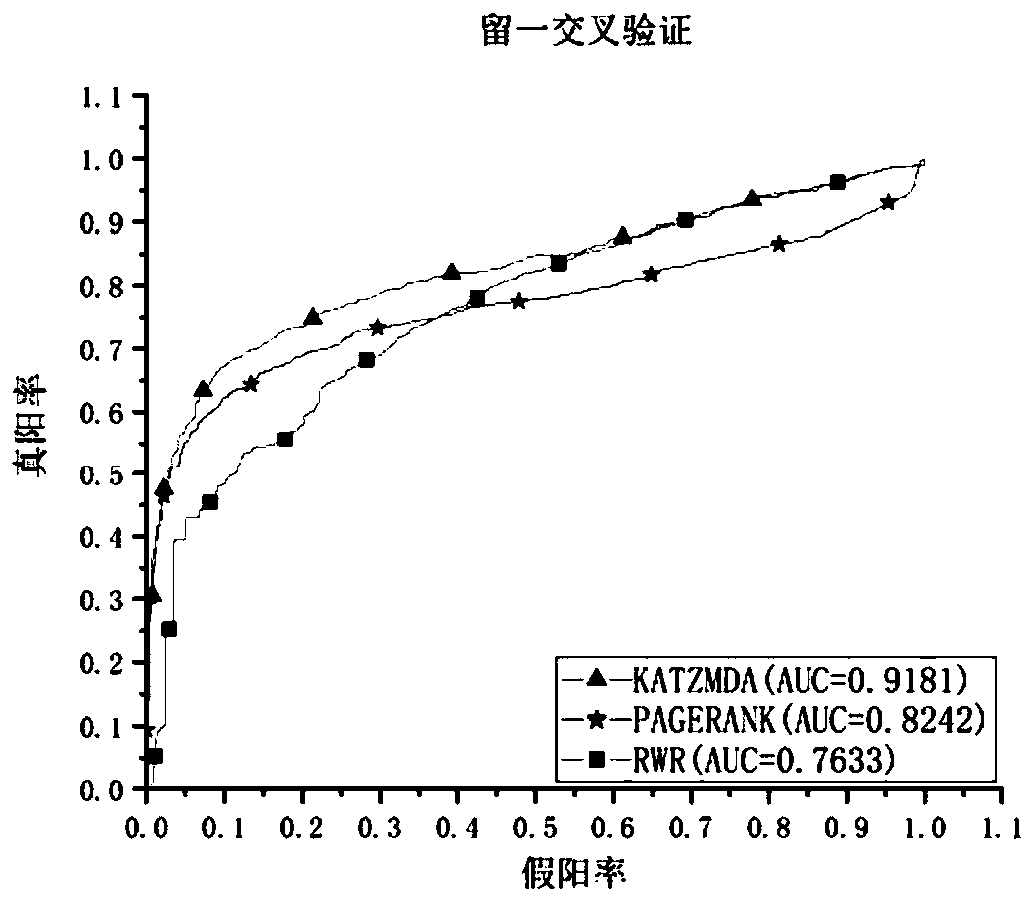
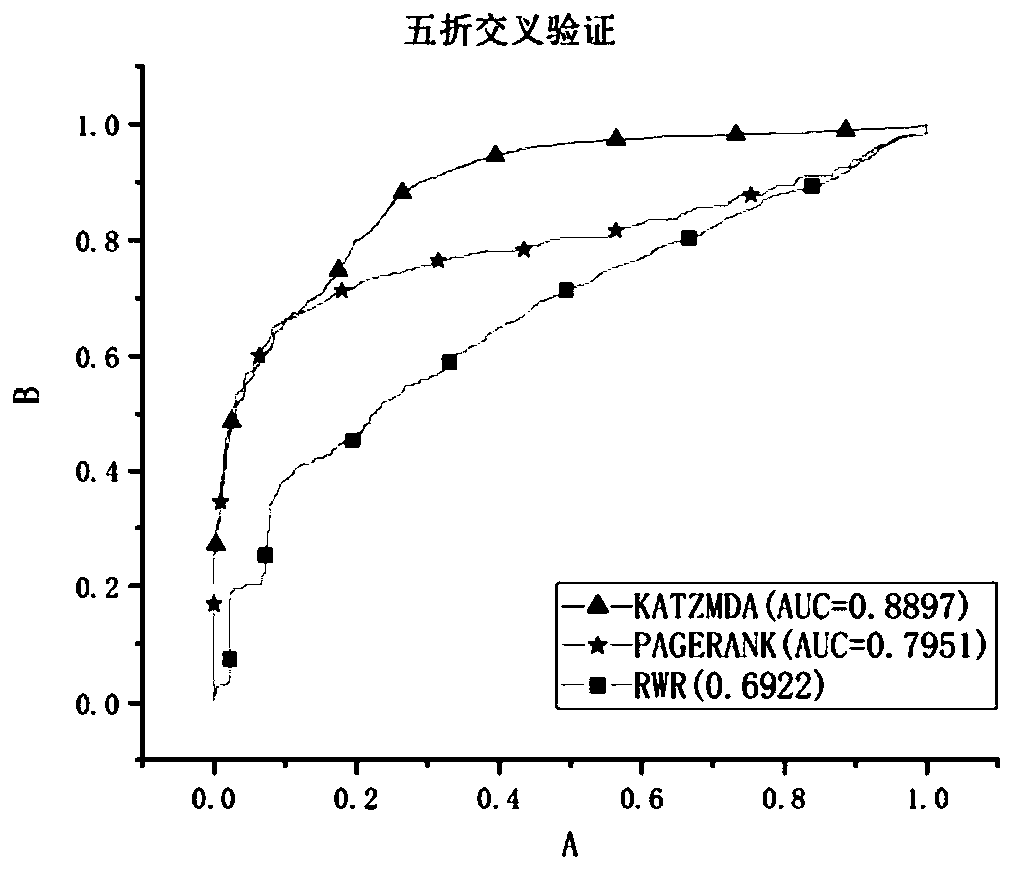
[0090] In this embodiment, the relevant metabolic data collected from the latest version of the HMDB database were screened and deduplicated, and 4537 known relationships were obtained, including 216 diseases and 2262 metabolites. Experiment platform is Windows 10 operating system, Intel (R) Core (TM) i5-8500CPU@3.00GHz processor, 8GB physical memory, realizes the method of the present invention with pycharm software.
[0091] 1. Transform the known relationship between metabolites and diseases extracted from the relevant metabolic data of the latest version of the HMDB database into known metabolites and disease networks:
[0092] First, the known relationship between metabolites and diseases is transformed into an adjacency matrix M(nd*nm), where nd represents the total number of disease...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com