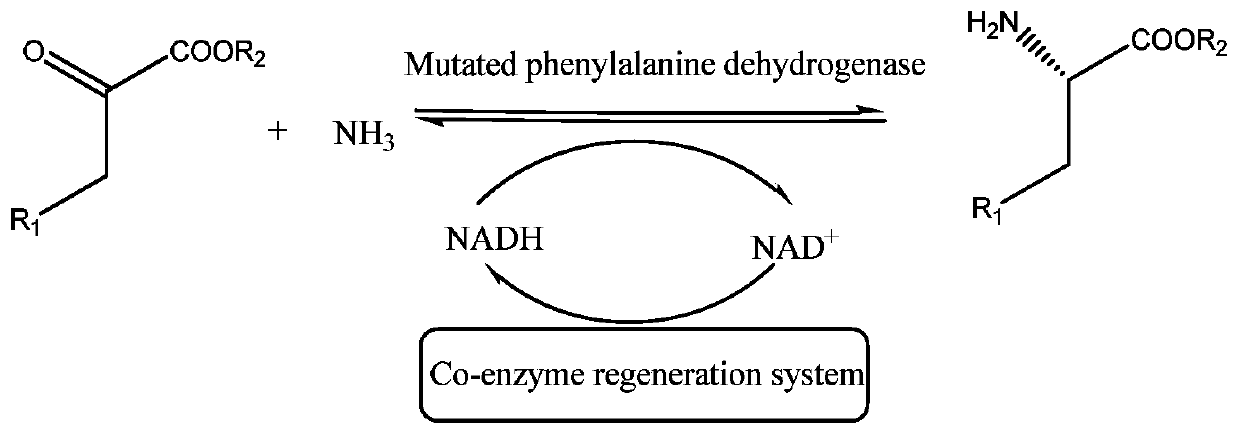
Phenylalanine dehydrogenase for catalytic preparation of non-natural amino acid and application of phenylalanine dehydrogenase
A technology of phenylalanine dehydrogenase and unnatural amino acid, applied in the field of molecular biology, can solve the problems of low catalytic activity of natural amino acid dehydrogenase, and achieve the effects of good industrial application prospect, convenient operation and high yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Embodiment 1: Construction of amino acid dehydrogenase mutant gene:
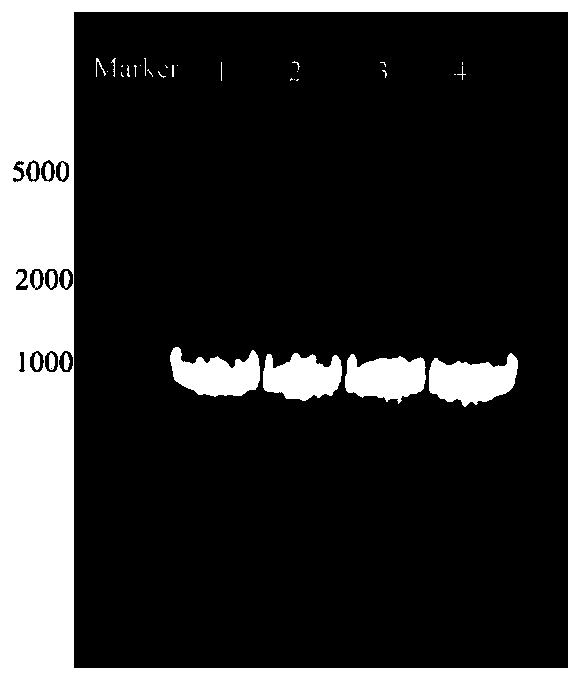
[0023] (1) In order to improve the catalytic activity of the amino acid dehydrogenase derived from Bacillus nanhaiensis (CGMCC NO.8969) (its amino acid sequence is SEQ D NO.07, and its encoding nucleotide sequence is SEQ ID NO.08), construct C - The amino acid sequence of the mutant with 4 amino acids reduced at the end is SEQ D NO.01, and the specific steps are as follows: use PCR to construct phenylalanine depletion sites with NdeI and Xhol restriction sites at the 5' end and 3' end respectively. Hydrogenase gene, PCR synthesis process was completed by Shanghai Sangon Bioengineering Technology Service Co., Ltd. like figure 1 As shown, after the PCR amplification product was identified by 1% agarose gel electrophoresis, the PheDH gene fragment was recovered from the gel, and double-digested with NdeI and XhoI enzymes, and the digested product was recovered, and the same double-digested pET-28a The ...
Embodiment 2
[0031] (1) In order to improve the activity of phenylalanine dehydrogenase derived from Bacillus nanhaiensis to catalyze unnatural amino acids, construct a mutant with 8 amino acids at the C-terminus, the sequence is shown in SEQ ID NO.02; the specific steps are as follows: The phenylalanine dehydrogenase gene with NdeI and Xhol restriction sites at the 5' end and 3' end was constructed by PCR method, and the PCR synthesis process was completed by Shanghai Sangon Bioengineering Technology Service Co., Ltd. like figure 1 As shown, after the PCR amplification product was identified by 1% agarose gel electrophoresis, the PheDH gene fragment was recovered from the gel, and double-digested with NdeI and XhoI enzymes, and the digested product was recovered, and the same double-digested pET-28a The plasmid (with His-tag tag) was ligated, and the ligated plasmid was transformed into Escherichia coli BL21(DE3) to obtain the pET28a-PheDH plasmid. The above plasmid was transformed into ...
Embodiment 3
[0036] (1) In order to improve the activity of phenylalanine dehydrogenase derived from Bacillus nanhaiensis to catalyze unnatural amino acids, construct a mutant with 12 amino acids at the C-terminus, the sequence is shown in SEQ ID NO.03; the specific steps are as follows:
[0037] The PCR method was used to construct the phenylalanine dehydrogenase gene with NdeI and Xhol restriction sites at the 5' end and 3' end respectively, and the PCR synthesis process was completed by Shanghai Sangon Bioengineering Technology Service Co., Ltd. like figure 1 As shown, after the PCR amplification product was identified by 1% agarose gel electrophoresis, the PheDH gene fragment was recovered from the gel, and double-digested with NdeI and XhoI enzymes, and the digested product was recovered, and the same double-digested pET-28a The plasmid (with His-tag tag) was ligated, and the ligated plasmid was transformed into Escherichia coli BL21(DE3) to obtain the pET28a-PheDH plasmid. The above...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com