Method for predicting health and immunity levels of introduced milk cows based on intestinal flora
A technology for intestinal flora and immunity, applied in the field of biotechnology, can solve the problems of long sequencing time, high sequencing cost, and complicated calculation, and achieve the effects of low cost, convenient sampling, and fast analysis speed.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
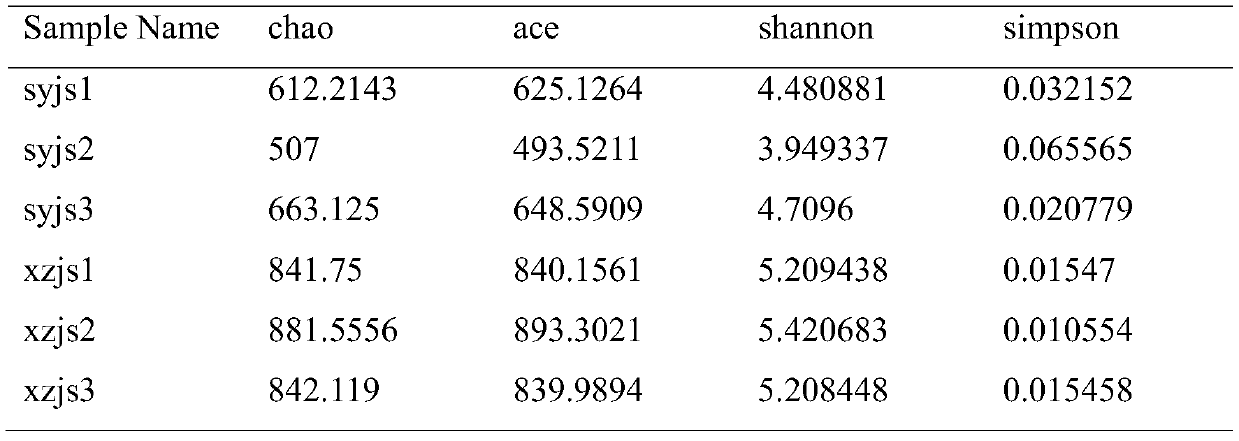
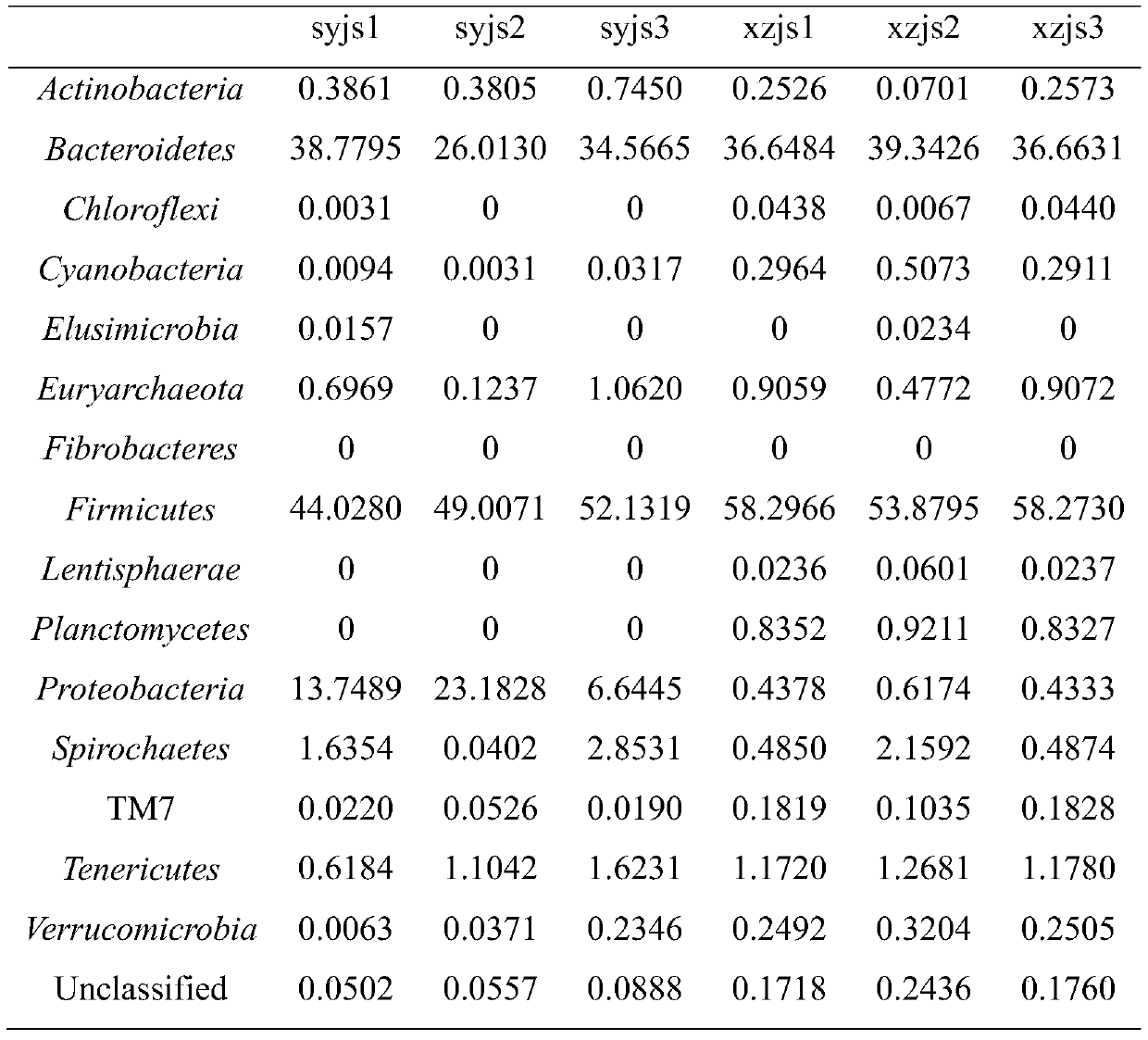
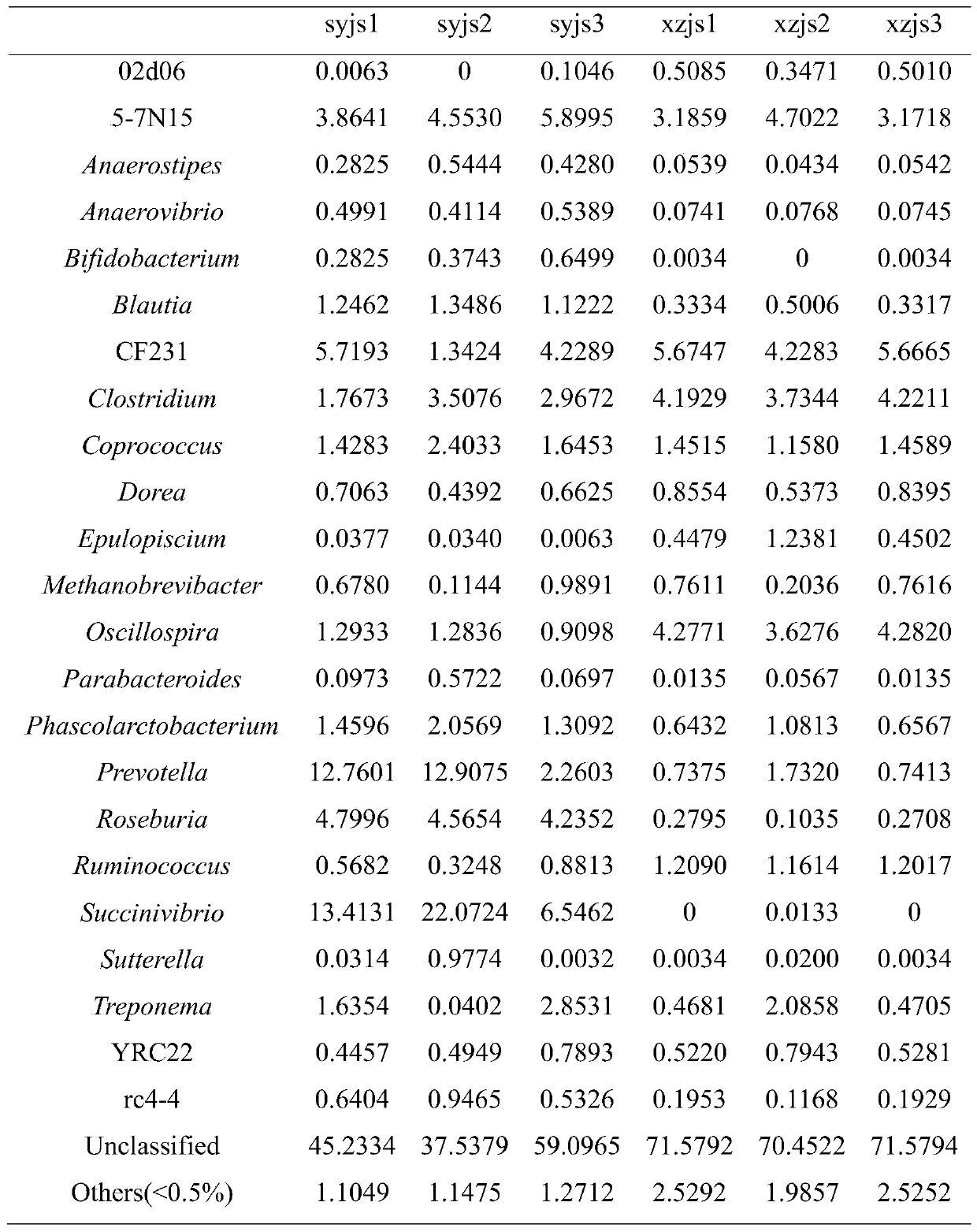
[0028] Step 1. Sample collection: After the farm makes a test request, it will receive relevant fecal sample collectors, one for each cow, to collect fecal samples from 3 cows that have been imported for less than half a year, numbered xzjs1, xzjs2, xzjs3, 3 respectively The feces samples of the first cows imported for more than half a year are numbered syjs1, syjs2, and syjs3 respectively, and the information on the month of introduction of each cow is recorded in detail. The feces are put into the collection tube containing the preservation solution and sent back to the sample center by courier. All samples were collected at room temperature and transported on dry ice.
[0029] Step 2. Establish database and variable region sequencing: use kit DNA purification technology to extract bacterial DNA in feces and establish database;
[0030] Sequencing fragments: The sequencing platform Illumima MiSeq sequencer sequenced the 16S variable region to obtain the original off-machine ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com