SNP molecular markers located on pig chromosome 16 related to pig lean meat percentage and eye muscle area and their application
An eye muscle area and molecular marker technology, applied in the field of molecular biotechnology and molecular markers, can solve the problems of little effect and long time, and achieve the effect of improving the breeding process, increasing the price, and speeding up the breeding process.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Embodiment 1 explains in detail the determination process that affects lean meat percentage in the present invention
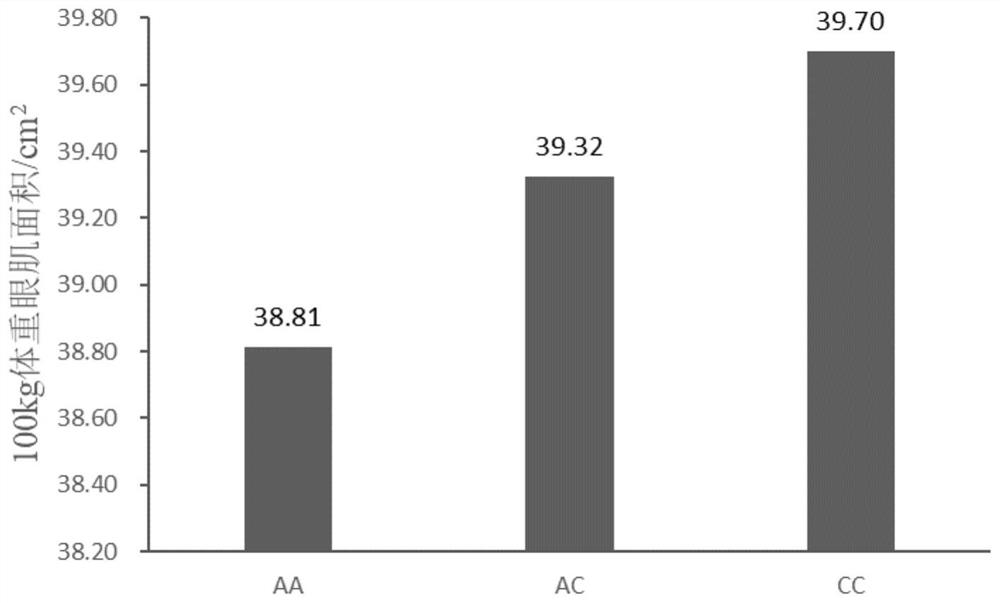
[0037] The lean meat percentage of live pigs with a body weight of 100kg was determined by the New Zealand HGS (Hennessy Grading System) carcass grading system. Aloka SSD 500V in vivo B-ultrasound measuring instrument was used to measure the eye muscle area of all test pigs when they were 100kg, and the measurement site was the 10th to 11th intercostal space. The measurement results are corrected by software for the area of the 100kg eye muscle. Use the standard measurement software package to calculate the eye muscle area, and directly print out the image and measurement value.
[0038] The experimental pig group used in the present invention is 2127 purebred plus-line Duroc of Guangdong Wen's Food Group Co., Ltd. Breeding Pig Branch Company, which is the core group of the Breeding Pig Branch Company, and the group pedigree records are detailed. ...
Embodiment 2
[0039] Embodiment 2 specifically explains the inventive process of obtaining the gene marker in the present invention
[0040] (1) The method of extracting DNA from the ear-like tissues of the Canadian Duroc pig was extracted from the whole genome DNA according to the marked phenol-chloroform method. Using Nanodrop-ND1000 spectrophotometer to detect the quality and concentration of the DNA of the purebred plus line Duroc population. The ratio of A260 / 280 is 1.8-2.0, and the ratio of A260 / 230 is 1.7-1.9. Finally, the qualified DNA samples were uniformly diluted to 50 ng / μl.
[0041] (2) 50K SNP genotype detection of the whole pig genome: GeneSeek Genomic Profiler Porcine50KSNP typing platform, using Illumina Infinium instructions and standard procedures for chip hybridization and result scanning. Finally, the genotype data were read by GenomeStudio software. The quality control of the obtained genotype data was carried out with PLINK v1.07, and the detection rate of eliminat...
Embodiment 3
[0047] Embodiment 3 specifically explains the inventive process of inventing and detecting SNP markers
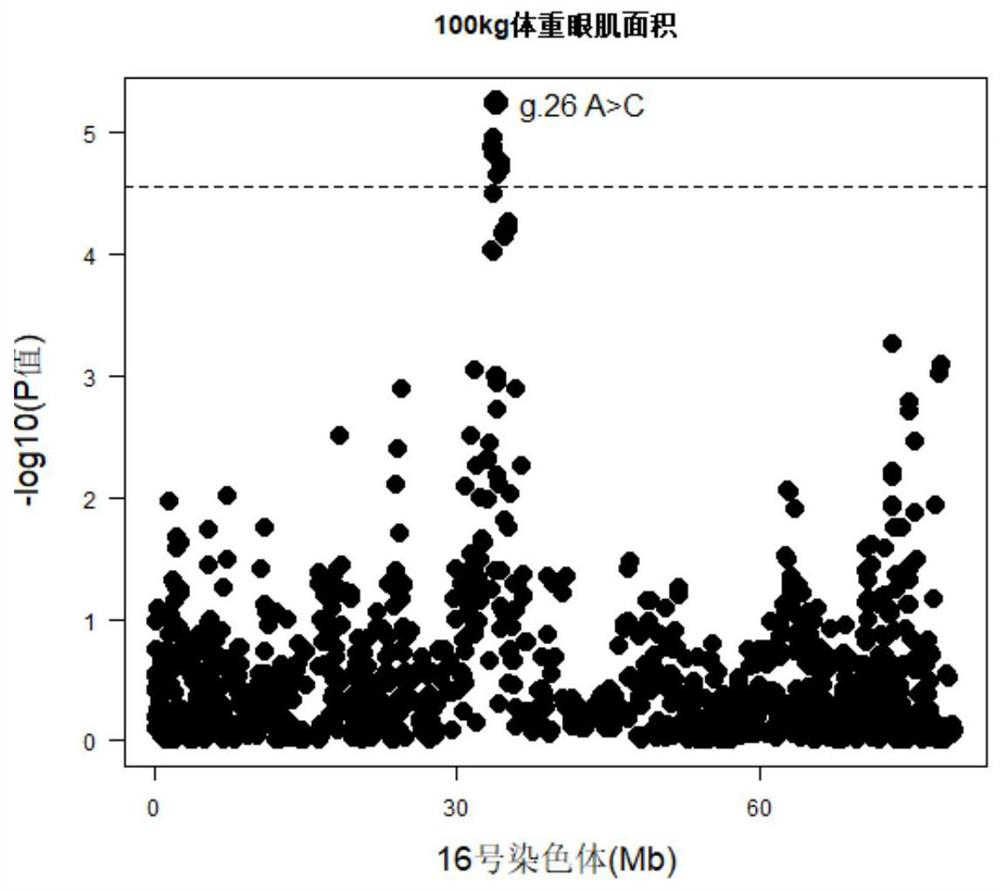
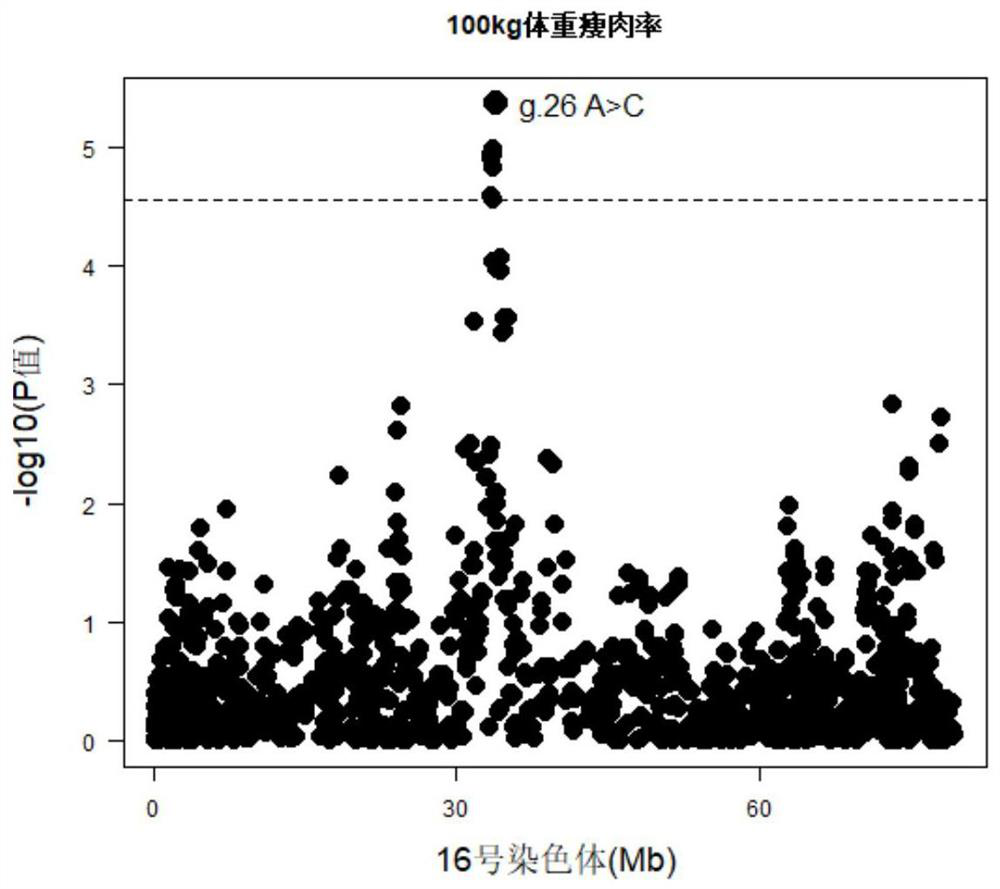
[0048] (1) The target segment containing the SNP site significantly related to the performance of the Canadian Duroc 100kg body weight eye muscle area and 100kg body weight lean meat rate is a 239bp nucleotide sequence in chromosome 16, and the upstream and downstream primers for sequence amplification It is primer-F and primer-R, and its nucleotide sequence is as follows:
[0049] Upstream primer primer-F: 5'-AGGTCAGAGTGGGGCAGTAA-3';
[0050] Downstream primer primer-R: 5'-AATGGGATTGGATGCGGGTT-3'.
[0051] (2) PCR amplification system and condition setting
[0052] Configure a 10 μL system, including 1 μL DNA sample, 0.3 μL upstream primer, 0.3 μL downstream primer, 5 μL PCR mix, ddH 2 O 3.4 μL, PCR conditions were pre-denaturation at 95°C for 5 min, denaturation at 95°C for 30 s, annealing at 64°C for 30 s, extension at 72°C for 30 s, a total of 35 cycles, and the fina...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com