Method for adjusting in-vitro migration of prostatic cancer cells through miR-203/SNAI2 axis
A mir-203, psin-mir-203 technology, applied in the field of molecular detection, can solve the problem that the interaction between miRNAs and SNAI2 has not been well elucidated
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0009] 1. Cell Culture
[0010] Five prostate cancer cell lines (DU145, PC3, 22Rv1, LNCaP, and C4-2) and two immortalized and untransformed prostate epithelial cell lines (PZ-HPV-7 and RWPE1) were obtained from ATCC, USA. Cells were cultured according to the standard manual of ATCC. Human umbilical vein endothelial cells (HuVECs) were purchased from Life Technologies (Carlsbad, California, USA) and cultured according to the instructions.
[0011] 2. Semi-quantitative RT-PCR and qRT-PCR
[0012] Semi-quantitative RT-PCR and qRT-PCR were performed as previously described. The primer sequences are as follows: 5´-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTAGTG-3´ (specific RT primer formiR-203), 5´-GTGCAGGGTCCGAGGT-3´ (real time PCR, miR-203, forward), 5´-GCCGCGTGAAATGTTTAGG-3´ (real time PCR , miR-203, reverse); 5´-CTCGCTTCGGCAGCACA-3´ (real time PCR, U6, forward), 5´-AACGCTTCACGAATTTGCGT-3´ (real time PCR, U6, reverse).
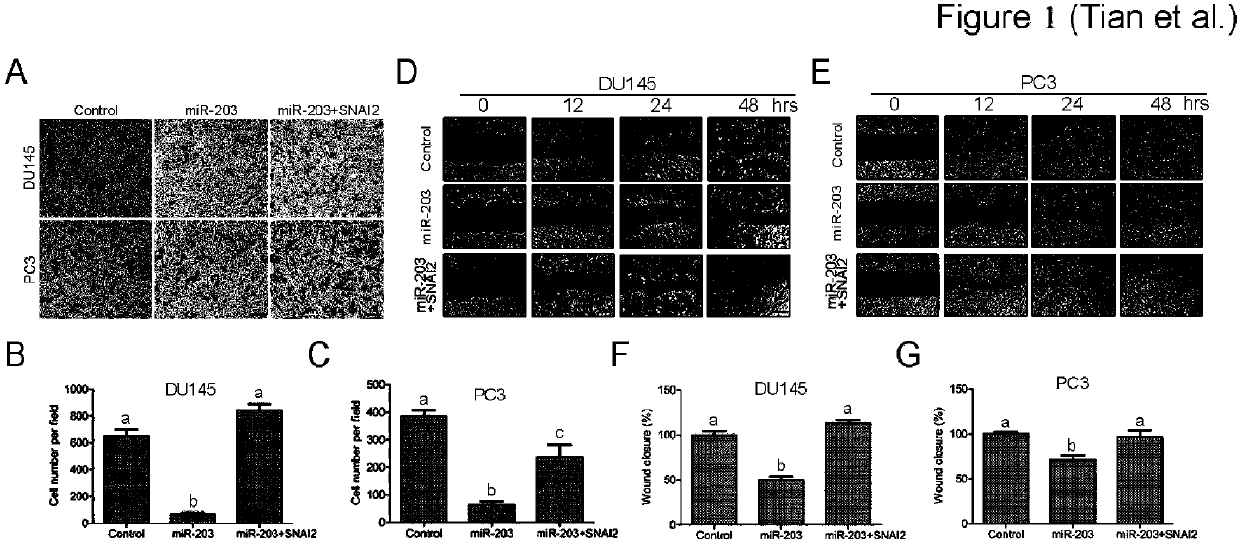
[0013] 3. Transwell infection detection
[0014] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com